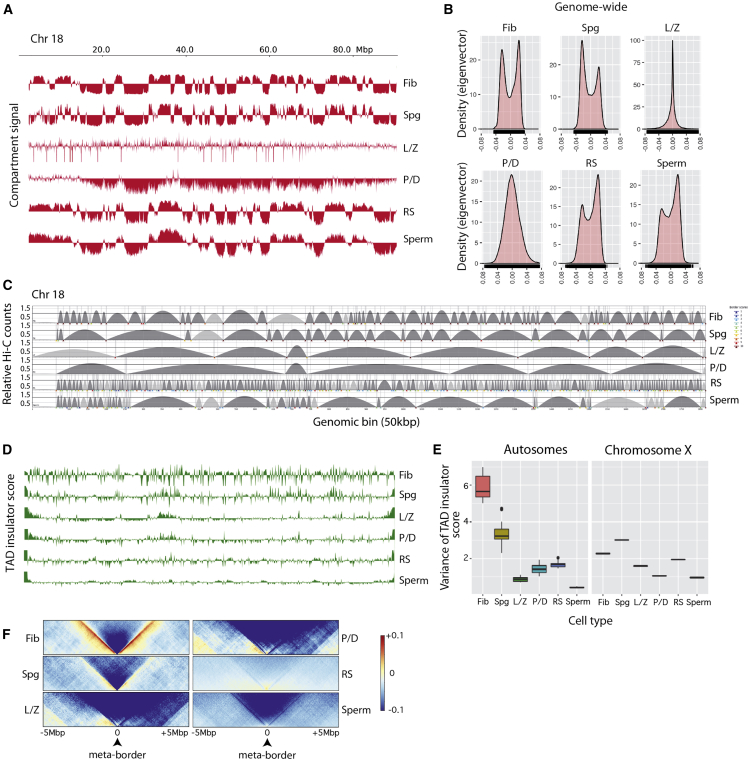
Figure 3.
Chromosome-specific A-B Compartment Profiles and TAD Signals
(A) Compartment signal (first eigenvector) across chromosome 18.
(B) Density plots of eigenvector values considering autosomes.
(C) TAD border alignments along chromosome 18. Dark gray arches represent TADs with higher intra-TAD interactions than expected. TAD border robustness (from 1 to 10) is represented by a color gradient.
(D) Representation of TAD insulator score in mouse chromosome 18.
(E) Variance of the TAD insulation scores for autosomes (left panel) and the X chromosome (right panel).
(F) Meta-plots for all TAD boundaries detected in fibroblasts (n = 2002), Spg (n = 834), L/Z (n = 305), P/D (n = 294), RSs (n = 4649), and sperm (n = 1042).
Fib, fibroblast; Spg, spermatogonia; L/Z, leptonema-zygonema; P/D, pachynema-diplonema; SpII, spermatocytes II; RS, round spermatids. See also Figures S3–S5.