Figure 7.
Higher-Order Chromatin Structure and Gene Expression in the X Chromosome
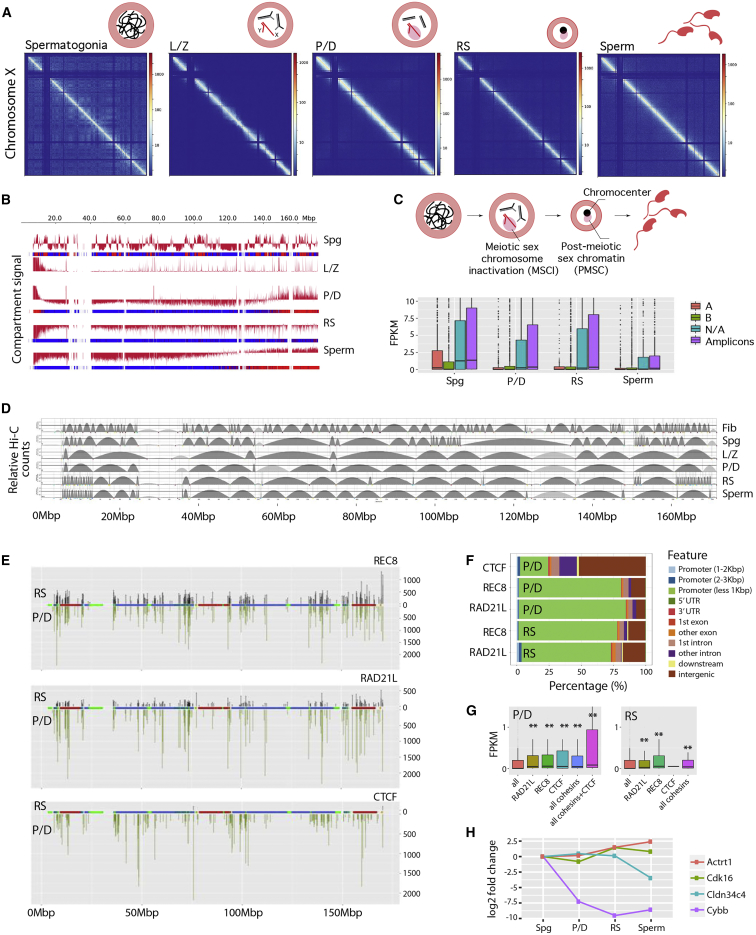
(A) ICE-corrected Hi-C matrices for the X chromosome in mouse germ cells, at a 50-Kbp resolution. Plaid blue regions correspond to non-mapped bins.
(B) Representation of compartment signal (first eigenvector) along the mouse’s X chromosome.
(C) Upper panel: Overview of spermatogenesis with two pairs of autosomes (black and gray lines) and the sex chromosomes (X and Y as red lines). Meiotic sex chromosome inactivation (MSCI) characterizes prophase I (shown as a pink cloud; also known as the sex body). In RSs, all centromeres associate in the center of the cell forming the chromocenter, with the X adjacent, forming the post-meiotic sex chromatin (PMSC). Lower panel: Boxplots of expression (FPKM values) of X genes binned according to A-B compartment or ampliconic. N/A are not assigned to a compartment or amplicon.
(D) X chromosome TAD alignments. Arches represent TADs with higher (darker gray) and lower (lighter gray) than expected intra-TAD interactions.
(E) CTCF and cohesin distribution on the X in P/D (green) and RSs (black). Ampliconic regions are green, and evolutionary strata are displayed, with blue representing older strata and red representing newer strata (see Figure S7 for further details).
(F) X chromosome distribution of CTCF and cohesins’ occupancy relative to TSSs and other genomic features in P/D and RS.
(G) Expression (FPKM values) of genes with CTCF and cohesin peaks located at promoter regions (−2 kbp from TSS). Asterisks represent statistically significant differential gene expression when compared with all genes in the mouse genome (p < 0.01). Boxes represent first and third quartiles, whereas black bars in boxes represent the median values.
(H) Expression changes (versus Spg) of representative X genes that reduce expression in P/D and maintain low levels during spermatogenesis (e.g., Cybb), genes that reduce expression in P/D and increase in RSs and sperm (i.e., Cdk16), genes that increase expression in RSs (e.g., Actr1), and genes that increase expression in P/D and then reduce (i.e., Cldn34c4).
Fib, fibroblast; Spg, spermatogonia; L/Z, leptonema-zygonema; P/D, pachynema-diplonema; RS, round spermatids. See also Figure S7.