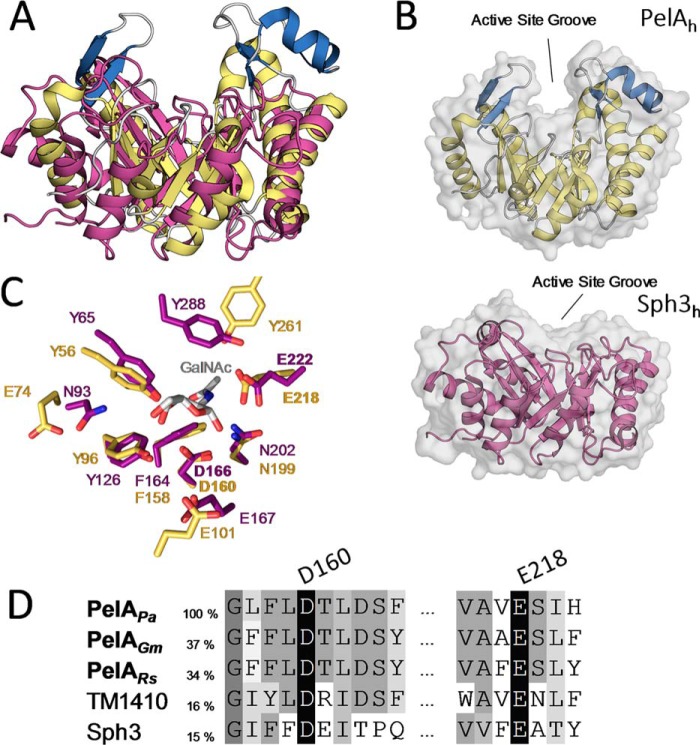
Figure 6.
PelAh and Sph3h differ in their substrate-binding cleft architecture but share catalytic motifs. A, tertiary structure alignment of Sph3h (PDB 5D5G, purple) with PelAh. B, transparent surface representation of PelAh (yellow and blue) and Sph3h (purple) in the same orientation shows the relative depths of the active site groove. C, alignment of the active site residues of PelAh (yellow) and Sph3h (purple) based on the Sph3h active site motifs shows high identity between the hydrolases around the GalNAc (gray)-binding site of Sph3h (PDB 5D6T). D, primary sequence alignment of P. aeruginosa PelAh (PelAPa) with homologues from Geobacter metallireducens (PelAGm), Ralstonia solanacearum (PelARs), as well as TM1410 and Sph3 (Aspergillus clavatus) done by MUSCLE. Sequence identity to PelAh is listed based on MUSCLE alignment for the two homologues and Sph3. Sequence identity to TM1410 is based on structural alignment.