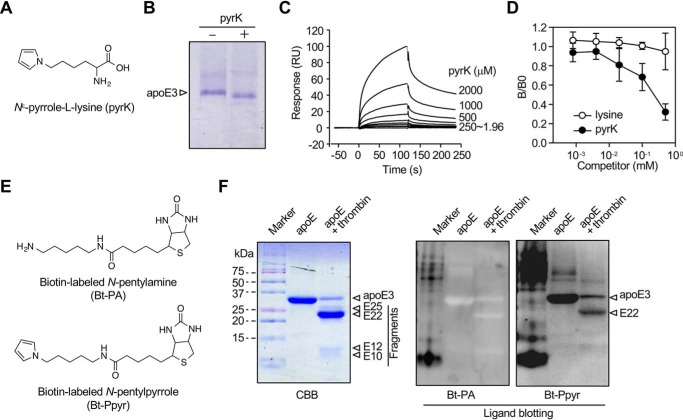
Figure 4.
Binding of apoE to pyrK. A, chemical structure of pyrK. B, mobility shift of apoE upon incubation with pyrK. C, surface plasmon resonance measurements. pyrK was subjected to an exchange reaction to preload it and tested for an interaction with apoE3 immobilized on a Biacore chip. Sensorgrams shows an increase in response units (RU) reflective of pyrK binding (association) and a slow decrease in response consistent with a loss of mass from washout (dissociation) after each injection (arrows). D, effect of pyrK on the binding of pyrrolated proteins to apoE3. apoE3 was preincubated by serial dilutions of pyrK before addition to the pyrrolated protein-coated ELISA plates. The experiment was performed in duplicate wells. Shown are the means ± S.D. of three independent experiments. E, chemical structures of biotin-labeled N-pentylamine (Bt-PA) and N-pentylpyrrole (Bt-Ppyr). F, ligand blot assay for the binding of biotin-labeled probes to thrombin-digested apoE. apoE was digested with thrombin for 3 h at 37 °C, and the resultant peptides were separated by 15% gel nonreducing SDS-PAGE. The gels were either stained with Coomassie Brilliant Blue R-250 (CBB) or transferred to PDVF membrane followed by ligand blotting with biotin-labeled N-pentylamine or biotin-labeled N-pentylpyrrole. The data are representative of three individual experiments.