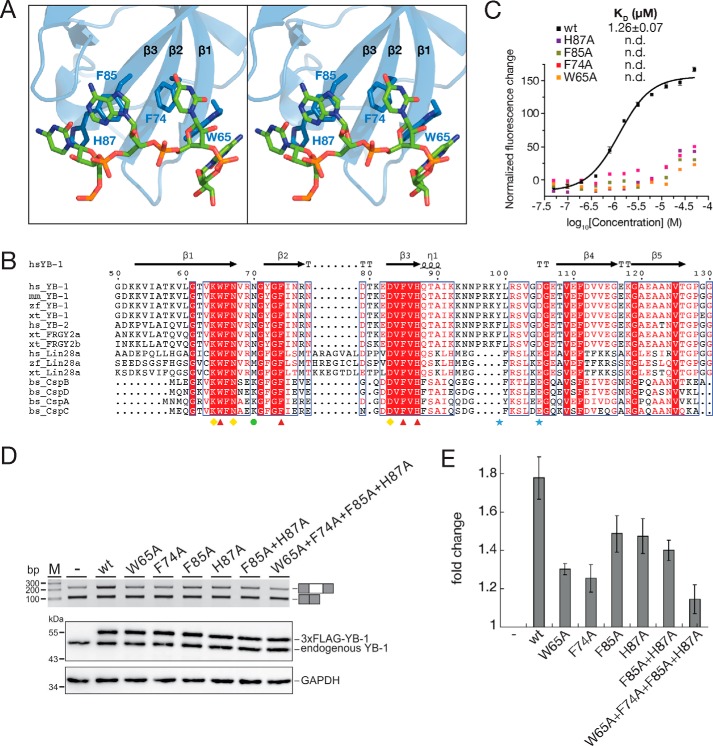
Figure 2.
π–π stacking interactions between YB-1 CSD and RNA oligo. A, stereo view of the interactions between YB-1 CSD and the RNA oligo (5′-U0C1A2U3C4U5-3′). B, sequence alignment of CSDs in cold shock proteins. Identical residues are indicated as white letters with red shading, and residues with similar properties are shown in red. Secondary structural elements are labeled above the sequences. Red triangles, aromatic residues involved in the π–π stacking interactions. Yellow diamonds, residues essential for the recognition. The green circle is specific for the 2′-hydroxyl group recognition of the fourth nucleotide. Blue stars, residues important for the YB-1 CSD dimerization. C, disruption of the π–π stacking interactions abolishes the binding between YB-1 CSD and RNA. The dissociation constants of the WT and mutant YB-1 CSD were measured by FP. D, top, RT-PCR analysis of in vivo splicing patterns of pET-v5 co-transfected with WT or mutant YB-1 constructs in HEK293 cells. Bottom, the expression level of YB-1 was detected by Western blotting, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. E, quantitation of D. The -fold changes of exon inclusion with S.D. values (error bars) were normalized to the exon inclusion percentage of pET-v5 (n = 3).