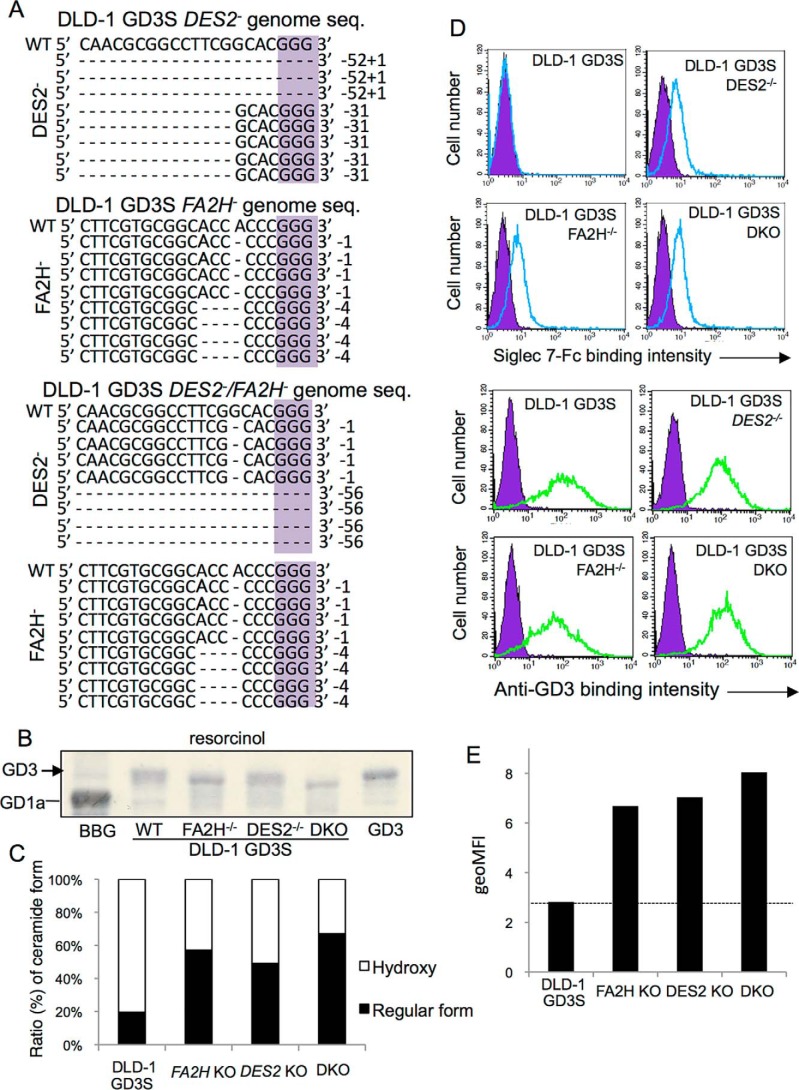
Figure 5.
Effects of knockout of DES2 and FA2H on the structure of GD3 and Siglec-7 binding. A, genome sequences of a total of eight clones each after knockout of DES2 and/or FA2H with CRISPR/Cas9 are shown. Because numbers of deleted bases were not in multiples of three in all clones, these gene products should lose correct protein forms. B, KO cell-derived ganglioside bands were compared in HPTLC. C, extracted acidic glycolipids from DLD-1 GD3S and individual KO cells were separated by HILIC–HPLC system and quantified by MRM analysis. The intensities of ions of major components of GD3 (t18:0–24:1/d18:1–24:0 h) and corresponding regular forms (d18:1–24:0) were measured and calculated as “hydroxyl” and “nonhydroxyl” (regular) species. D, FCM analysis by Siglec-7–Fc (upper panel) and anti-GD3 mAb (lower panel). E, summary of FCM analysis. Geometric mean of fluorescence intensity (geoMFI) of Siglec-7-binding was measured and presented. These analyses were repeated at least three times with similar results.