Figure 2:
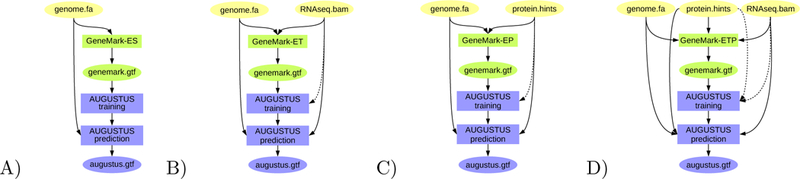
BRAKER pipeline: A) Training GeneMark-ES on genome data only; ab initio gene prediction with AUGUSTUS. B) Training GeneMark-ET supported by RNA-Seq spliced alignment information, prediction with AUGUSTUS with that same spliced alignment information. C) Training GeneMark-EP on protein spliced alignment information, prediction with AUGUSTUS with that same spliced alignment information. D) Training GeneMark-ETP supported by RNA-Seq alignment information and protein spliced alignment information, prediction with AUGUSTUS using the same alignment information. Introns supported by both RNA-Seq and protein alignment information are trusted - their prediction in gene structures by GeneMark-ETP and AUGUSTUS is enforced. Proteins used for C) and D) can be of longer evolutionary distance.
