Figure 3:
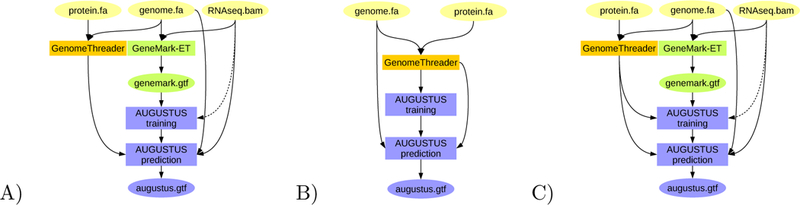
BRAKER pipelines for integration of protein information from closely related species: A) Training GeneMark-ET supported by RNA-Seq spliced alignment information, prediction with AUGUSTUS with spliced alignment information from RNA-Seq data and with gene features determined by alignments from proteins of a very closely related species against the target genome. B) Training AUGUSTUS on the basis of spliced alignment information from proteins of a very closely related species against the target genome. C) Training GeneMark-ET on the basis of RNA-Seq spliced alignment information, training AUGUSTUS on a set of training gene structures compiled from RNA-Seq supported gene structures predicted by GeneMark-ET and spliced alignment of proteins of a very closely related species.
