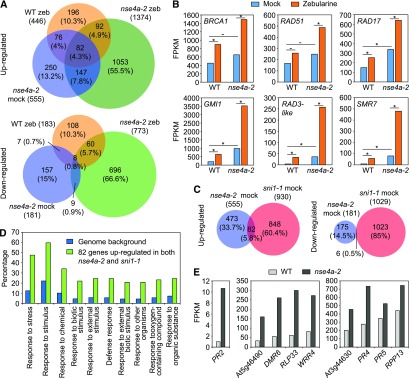
Figure 6.
Transcriptome Analysis of nse4a-2 Plants.
(A) Venn diagrams of genes significantly (DESeq, adjusted P < 0.05) up- and downregulated in dissected shoot apices of the 20 µM zebularine (zeb)–treated wild-type (WT zeb/WT mock), mock-treated nse4a-2 (nse4a-2 mock/WT mock), and 20 µM zeb-treated nse4a-2 (nse4a-2 zeb/nse4a-2 mock) plants. The data are based on two RNA sequencing replicates.
(B) mRNA abundance of DNA damage repair marker genes expressed as fragments per kilobase per million of reads (FPKM) based on data shown in (A). Asterisks and dashes indicate statistically significant and nonsignificant, respectively, differences between groups indicated by horizontal bar in DESeq (adjusted P-value < 0.05). WT, wild type; BRCA1, BREAST CANCER SUSCEPTIBILITY1; RAD51, RADIATION SENSITIVE51; RAD17, RADIATION SENSITIVE17; GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED1; RAD3-like,RADIATION SENSITIVE3-like, At1g20750; SMR7, SIAMESE-RELATED7.
(C) Venn diagrams of significantly up- and downregulated genes in nse4a-2 (see [A]) and sni1-1 (sni1-1 mock/wild type (WT) mock; ATH1 expression microarrays, adjusted P < 0.05) plants.
(D) Gene ontology (GO) term analysis of 82 genes significantly upregulated in both nse4a-2 and sni1-1 (see [C]) using agriGO v2.0. Top 10 GO term categories are shown as input relative to Arabidopsis genomic background/reference. The full list of significant GO terms is available in Supplemental Table 3.
(E) Examples of significantly (DESeq, adjusted P-value < 0.05) upregulated defense-related genes in dissected shoot apices of mock-treated nse4a-2 plants. DMR6, DOWNY MILDEW RESISTANT6; RLP33, RECEPTOR LIKE PROTEIN33; WRR4, WHITE RUST RESISTANCE4; RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13.