Fig. 4.
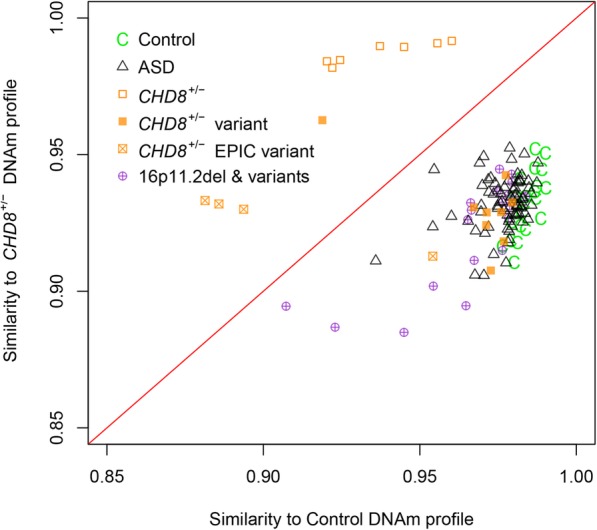
Evaluating the sensitivity of the CHD8+/− DNAm signature using additional CHD8 test cases (EPIC array). Additional independent test cases consisting of CHD8 sequence variants (n = 4; orange squares with cross) were run on the EPIC array. Of the 103 sites in the 450K-derived CHD8+/− DNAm classification signature, 92 sites overlapped those of the EPIC array and were thus used to classify the additional EPIC test cases. One VUS case received a negative classification score. Three of the four additional CHD8 test cases were accurately classified as more similar to CHD8+/− signature training cases (open orange square); these cases were known to have pathogenic CHD8 variants, demonstrating 100% sensitivity. The one sample classified as benign was an inherited missense variant, similar to those 450K test cases receiving negative scores. All other cases and controls (450K) were accurately classified as in Fig. 3
