Figure 1. Domain structure of Epacs and mechanism of activation of Epac2.
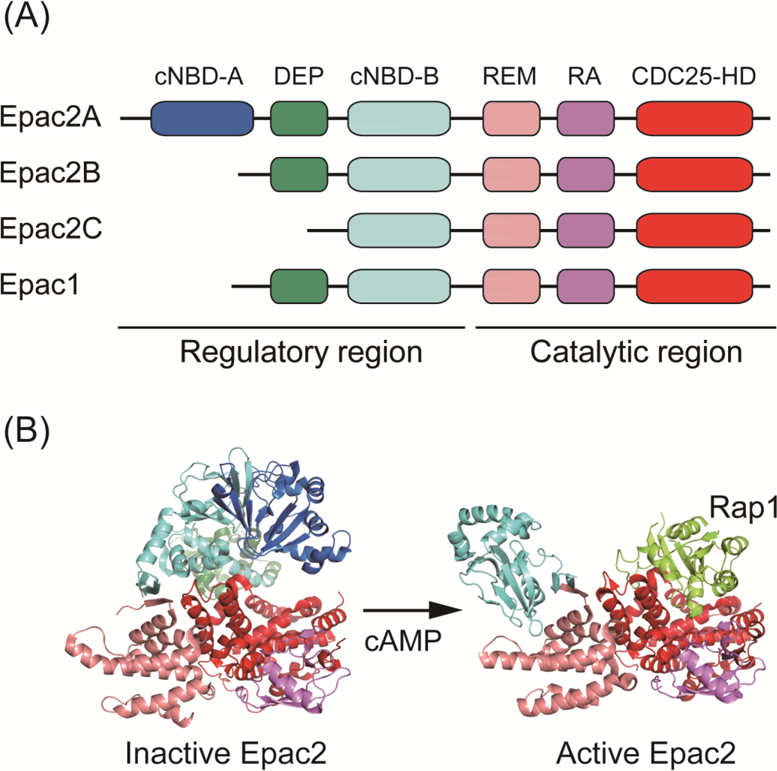
(A) Domain structure of Epac1 and Epac2. Epacs have an amino-terminal regulatory region and a carboxy-terminal catalytic region. The regulatory region contains one or two cyclic nucleotide binding domains (cNBD) and a DEP (Dishevelled, Egl-10, and Pleckstrin) domain. The catalytic region contains a REM (Ras exchange motif), a RA (Ras association) domain and a CDC25-HD (CDC25 homology domain).
(B) Crystal structures of inactive-form (PDB code 2BYV) and active-form (in complex with Rap1B and cAMP analog; PDB code 3CF6) of Epac2. In the inactive-form of Epac2, the Ras-binding region in CDC25-HD is masked by the regulatory region. The binding of cAMP to cNBD-B induces a conformational change, enabling access by Rap to the catalytic region.
