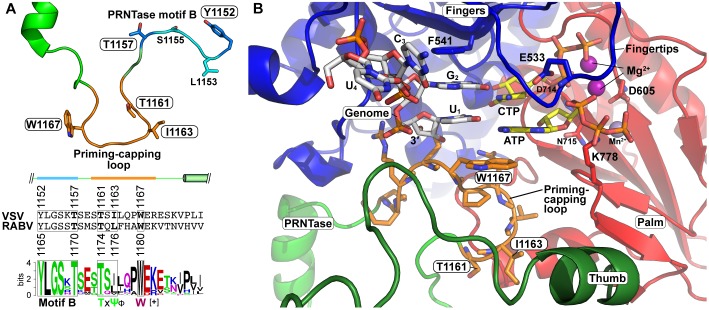
FIGURE 10.
Formation of the VSV terminal de novo initiation complex. (A) The three-dimensional structural model of the priming-capping loop (residues 1160–1169, orange), with its flanking regions including PRNTase motif B, of the VSV L protein is represented as a ribbon diagram with stick models of key amino acid residues (Ogino et al., 2019) (upper). An amino acid sequence logo for putative priming-capping loops and their franking sequences of 110 vertebrate and arthropod rhabdoviruses (Ogino et al., 2019) are shown with the corresponding sequences of VSV and RABV (lower). The secondary structures of this region in the VSV L protein are depicted above its sequence. For amino acid symbols, see Figure 5A. (B) The structure of the VSV L protein in complex with the 3′-terminal sequence (3′-UGCU-5′) of the genome (white carbon backbone), initial (ATP) and incoming (CTP) nucleotides (yellow carbon backbone), two Mg2+ ions (purple), and Mn2+ ion (obscured) was modeled as described in Ogino et al. (2019) (Model Archive id: ma-5k432). W1167 on the priming-capping loop (orange carbon backbone) π-stacks with the initiator ATP. Key amino acid residues are shown as stick models on the fingers and palm subdomains. The RdRp subdomains and PRNTase/priming loop are colored as in Figure 5.