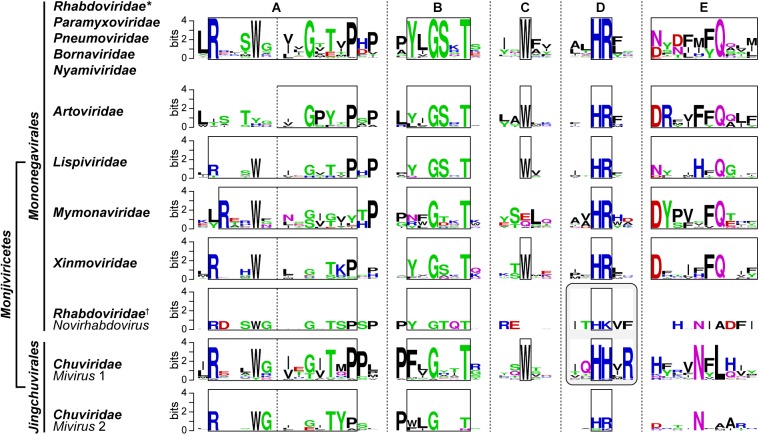
FIGURE 12.
Conservation of PRNTase motifs in L proteins of NNS RNA viruses and their related viruses. Local amino acid sequences (PRNTase motifs A–E) of PRNTase-like domains in L proteins of 7 peropuviruses (Artoviridae), 6 arliviruses (Lispiviridae), 10 sclerotimonaviruses (Mymonaviridae), 8 anpheviruses (Xinmoviridae), and 4 novirhabdoviruses (Rhabdoviridae†) belonging to the order Mononegavirales and 29 miviruses [group 1, 23 viruses with a circular genome(s); group 2, 6 viruses with a linear genome, Chuviridae] belonging to the order Jingchuvirales were analyzed by the WebLogo program (Crooks et al., 2004). The resulting sequence logos are shown with those for NNS RNA viruses belonging to the Rhabdoviridae (∗, except novirhabdoviruses), Paramyxoviridae, Filoviridae, Bornaviridae, and Nyamiviridae families (top, as in Figure 8A).