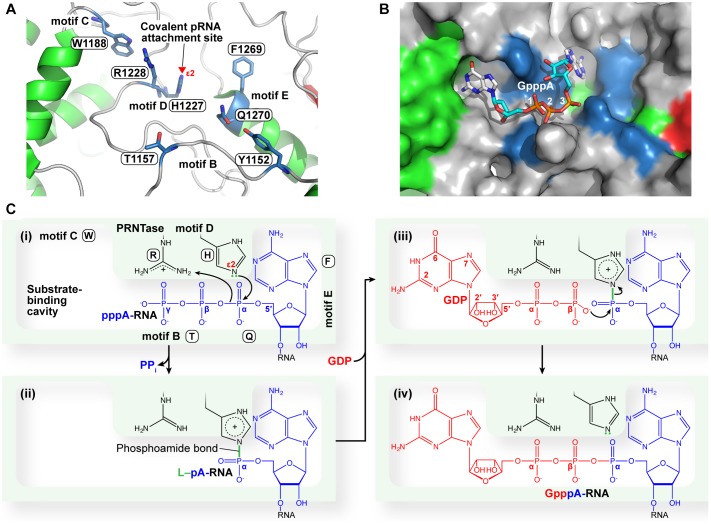
FIGURE 9.
A proposed catalytic mechanism of mRNA capping by the PRNTase domain of the VSV L protein. (A) The three-dimensional structure of the VSV PRNTase active site (PDB id: 5A22) is shown as a ribbon diagram, in which α-helices, β-sheets, and loops are colored green, red, and gray, respectively. The key amino acid residues in PRNTase motifs B–E (Neubauer et al., 2016) are shown as stick models (blue carbon backbone). The covalent pRNA attachment site (Nε2 atom) of catalytic H1227 in motif D (Ogino et al., 2010) is indicated by a red arrowhead. The side-chain carbonyl group of Q1270 in motif E interacts with the hydroxyl group of Y1152 in motif B via hydrogen-bonding. (B) A surface representation of the view in (A) is shown with a docked GpppA 5′-cap analog in stick model. The GpppA resides in a crevice heavily generated by motifs B–E. Docking was performed with Autodock Vina (Trott and Olson, 2010). (C) The 5′-pppA residue of VSV pre-mRNA may dock into one side of a putative substrate-binding cavity surrounded by motifs E, D, and B (i), where a nucleophile formed on the Nε2 position of H1227 subsequently attacks the α-phosphorus in the 5′-triphosphate group of pre-mRNA to form the covalent L–pRNA intermediate (ii). GDP may dock into another side of the putative substrate-binding pocket surrounded by motifs C, D, and B (iii). There, an oxyanion on the β-phosphate of GDP nucleophilically attacks the α-phosphorus of pRNA linked to H1227, resulting in the formation of the GpppA cap structure on pre-mRNA (iv).