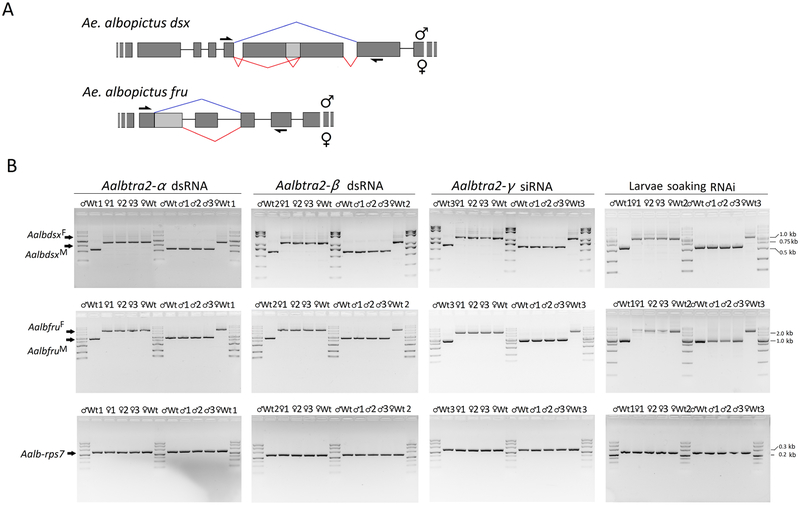
Figure 6. Sex-specific splicing patterns of the dsx and fru transcripts in Aedes albopictus adults after individual or triple RNAi knockdown of Aalbtra2-α, Aalbtra2-β, and Aalbtra2-γ.
(A) Genomic organization and splice variants of dsx and fru in Ae. albopictus. The male-specific and female-specific splicing patterns are marked in blue and red, respectively. The exons and introns are not drawn to scale (see Supplementary Table 3 for further details). The arrows denote the locations of the primers used to amplify the cDNA products of dsx and fru. The primer sets encompassed the alternative splicing junctions and thus produced sex-specific RT–PCR products. (B) RT–PCR products obtained using mRNA from individuals 48 hr after injection of siRNAs/dsRNA targeting one of the three tra2 genes (columns 1–3) or from adult individuals in which all Aalbtra2 genes had been knocked down since the larval stage through soaking with Aalbtra2-α and Aalbtra2-β dsRNA and Aalbtra2-γ siRNA mixtures (column 4).♂1–3 and ♀1–3 refer to the three replicate groups (n=15 adults per replicate) of tra2 RNAi males and females, respectively. Wt♂1–3 and Wt♀1–3 indicate the three replicate groups (n=15 per replicate) of wild-type males and females. Fragments corresponding to the fru female (AalbfruF; 2010 bp) and male (AalbfruM; 987 bp) cDNA sequences and the dsx female (AalbdsxF; 1062 bp) and male (AalbdsxM; 620 bp) cDNA sequences were resolved by electrophoresis on a 1.5% agarose gel that was stained with ethidium bromide and photographed. The numbers with short lines on the right side indicate the molecular weights. Ae. albopictus ribosomal protein 7 (rpS7) mRNA was used as an internal control.