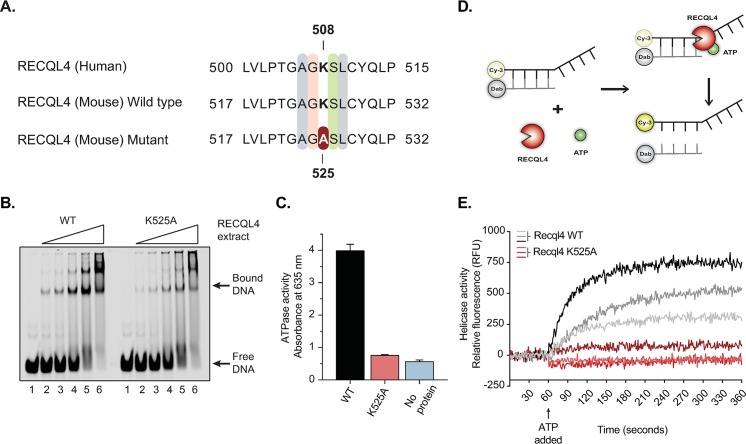
Fig 1. The Recql4K525A mutation results in a biochemically inactive helicase protein.
(A) Sequence homology between human and mouse RECQL4 showing the highly conserved amino acids between species. The helicase-dead mutation was achieved by replacing lysine by alanine in the 525 position in the mouse (508 in humans). (B) Electrophoretic-mobility shift assay (EMSA) comparing the DNA binding of WT versus K525A mutant Recql4. Lanes 1–6 show serial dilutions of Recql4 protein ranging from 19nM to 300nM bound to 25nM ssDNA oligo XOm1 conjugated to IRDye680. (C) In vitro ATPase assay of WT versus K525A mutant was obtained by measuring the absorbance at 635 nm. Proteins were assayed at 115nM in the presence of 1mM ATP and DNA in assay buffer. (D) Schematic representation of the fluorescence-based helicase assay. The dsDNA substrate with a ssDNA loading site is attached to a fluorescent donor (Cy-3) and a quencher acceptor (Dab). In the presence of ATP, the helicase (Recql4) is activated and separates the two complementary strands releasing the fluorescent probe. (E) Helicase assay of WT versus K525A mutant. After recording baseline fluorescence for 60 seconds, ATP was added, and helicase activity was measured in relative fluorescence units (RFU) for three different concentrations of Recql4 protein.