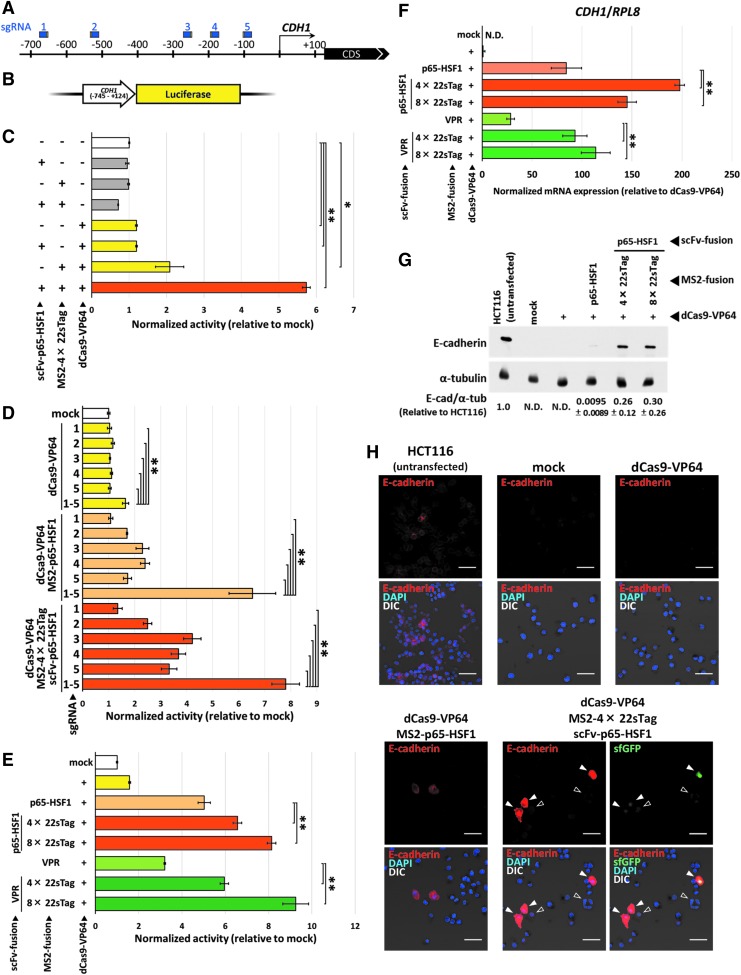
FIG. 2.
Activation of CDH1 in MIA-PaCa2 cells with multiplex TREE system. (A) Schematic illustration of the positions of sgRNAs used for the activation of CDH1. Protospacer and protospacer adjacent motif (PAM) sequences are shown as blue and yellow boxes, respectively. See also Supplementary Figure S2A. (B) Schematic illustration of luciferase reporter vector containing CDH1 promoter and 5stratio (C) Initial validation of the TREE system by reporter assay. All possible patterns of non-, single, double, and triple administration of the TREE components were tested. Data are shown as the mean ± standard deviation (SD; n = 4). **p < 0.01; *p < 0.05. (D) Activity comparison among single and five sgRNAs, using the dCas9-VP64, SAM, and TREE systems by the reporter assay. Data are shown as the mean ± SD (n = 4). **p < 0.01. (E) Activity comparison among the effectors and the repeat numbers of GCN4 epitopes, as well as the MS2-effector system, by reporter assay. Data are shown as the mean ± SD (n = 4). **p < 0.01; *p < 0.05. (F) Endogenous CDH1 expression quantified by quantitative polymerase chain reaction (qPCR). Data are shown as the mean ± SD (n = 4). **p < 0.01; *p < 0.05. N.D., not detected. (G) Detection of E-cadherin and α-tubulin proteins by immunoblotting. Loaded protein mass is as follows: for the detection of E-cadherin in HCT116 and α-tubulin in all samples, 3 μg; for the detection of E-cadherin in all samples other than HCT116, 10 μg. Data are shown as the mean ± SD (n = 3), and one set of blots is shown. N.D., not detected. The other blots are shown in Supplementary Figure S3A. (H) Detection of E-cadherin protein by immunostaining. Filled and open triangles indicate E-cadherin/sfGFP- and sfGFP-positive cells, respectively. Scale bars, 50 μm. See also Supplementary Figure S3B.