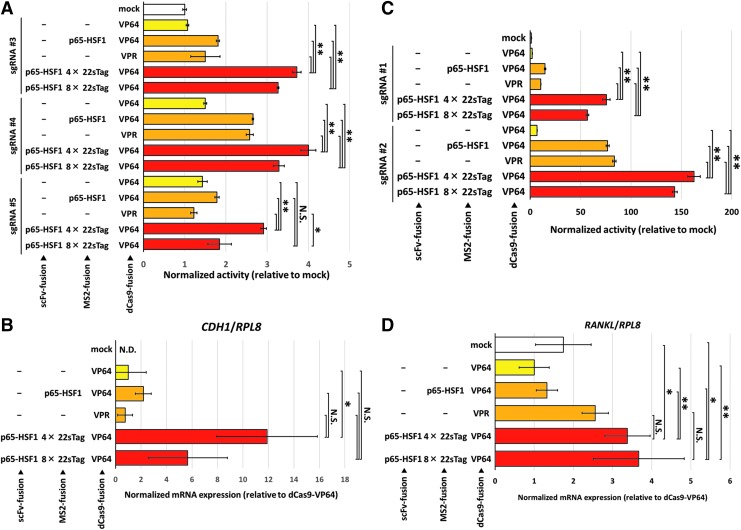
FIG. 4.
Robust transcriptional activation with single TREE system. (A) Assessment of the activation capacity of single TREE systems containing sgRNA #3, #4, or #5 by CDH1 reporter assay in MIA-PaCa2 cells with the comparison with the previous systems. Data are shown as the mean ± SD (n = 4). **p < 0.01; *p < 0.05. N.S., not significant. (B) Assessment of the activation capacity of single TREE systems containing sgRNA #5 by qPCR analysis of endogenous CDH1 gene in MIA-PaCa2 cells with the comparison with the previous systems. Data are shown as the mean ± SD (n = 3). *p < 0.05. N.S., not significant; N.D., not detected. (C) Assessment of the activation capacity of single TREE systems containing sgRNA #1 or #2 by RANKL reporter assay in HEK293T cells with the comparison with the previous systems. Data are shown as the mean ± SD (n = 4). **p < 0.01. (D) Assessment of the activation capacity of single TREE systems containing sgRNA #2 by qPCR analysis of endogenous RANKL gene in HEK293T cells with the comparison with the previous systems. Data are shown as the mean ± SD (n = 4). **p < 0.01; *p < 0.05. N.S., not significant. For dCas9-VPR, sgRNAs without MS2 stem loops were used instead of sgRNA2.0.