Fig. 4. Impact of the viral RNA on filament stability and structure of CP.
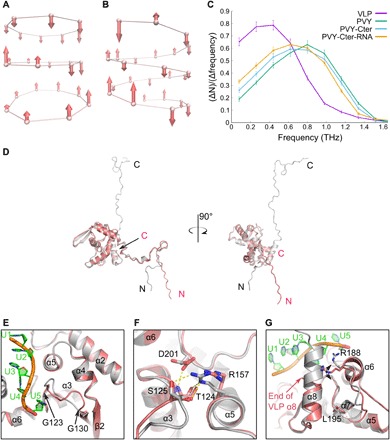
(A to C) Molecular dynamics of filaments. Axial displacements of protein unit centers of mass for the lowest vibrational modes: (A) VLP (ω = 0.0066 THz) and (B) PVY (ω = 0.0444 THz), as calculated by PCA (principal components analysis). Each protein unit is represented by a center of mass of the Val44-Ile218 region. (C) Frequency distributions of protein units’ vibrations along the fibril axis for different structural models: VLP, PVY, PVY without the C-terminal region (PVY-Cter), and PVY without C-terminal region and ssRNA (PVY-Cter-RNA). Error bars represent scattering in different time sections of the trajectory. (D) Superposition of PVY CP (gray) and VLP CP (pink) on core regions, shown at two angles. (E to G) Structural differences in CP core regions of PVY and VLP; the color code is as in (A). (E) The loop between Gly123 and Gly130. (F) Highly conserved residues Ser125, Arg157, and Asp201 involved in PVY ssRNA binding. Electrostatic interactions are shown as dashed lines. For clarity, the C-terminal region and ssRNA were removed from the picture. (G) Residues Arg188-Leu195 and α8 helix. The black arrow indicates flexibility of Arg188 in VLP in comparison to the fixed Arg188 in PVY CP.
