Fig. 3.
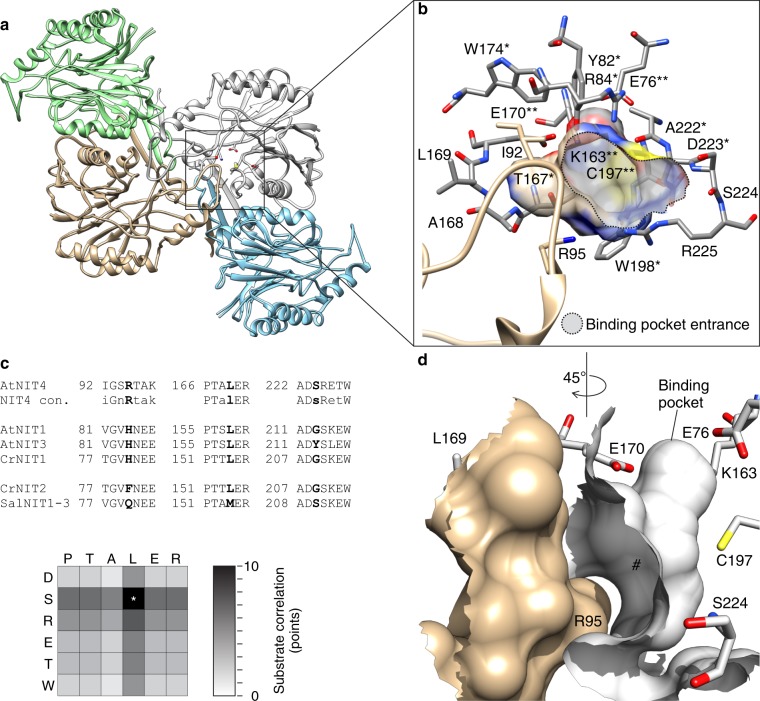
The substrate-binding pocket. a AtNIT4 structure showing two dimers interacting across the C-interface, the position of the catalytic site is indicated by a square. b The active-site pocket, the catalytic residues (E76, K163, E170, and C197) are shown (**), residues conserved in all plant NITs (*). A loop (tan) arising from the adjacent monomer (between α2 and α3) extends over the entrance to the active-site pocket. c As expected, the amino acid residues surrounding the binding pocket correlate with substrate specificity. Three amino acid positions in particular are responsible for the majority of the variation between known plant NITs: 169, 224, and 95 (numbered according to AtNIT4). d A lid (tan), formed by the loop, limits the length of the binding pocket (#). R95 is in the lid loop; S224 is on the outer border of the binding pocket and L169 points toward the light green subunit across the C-interface