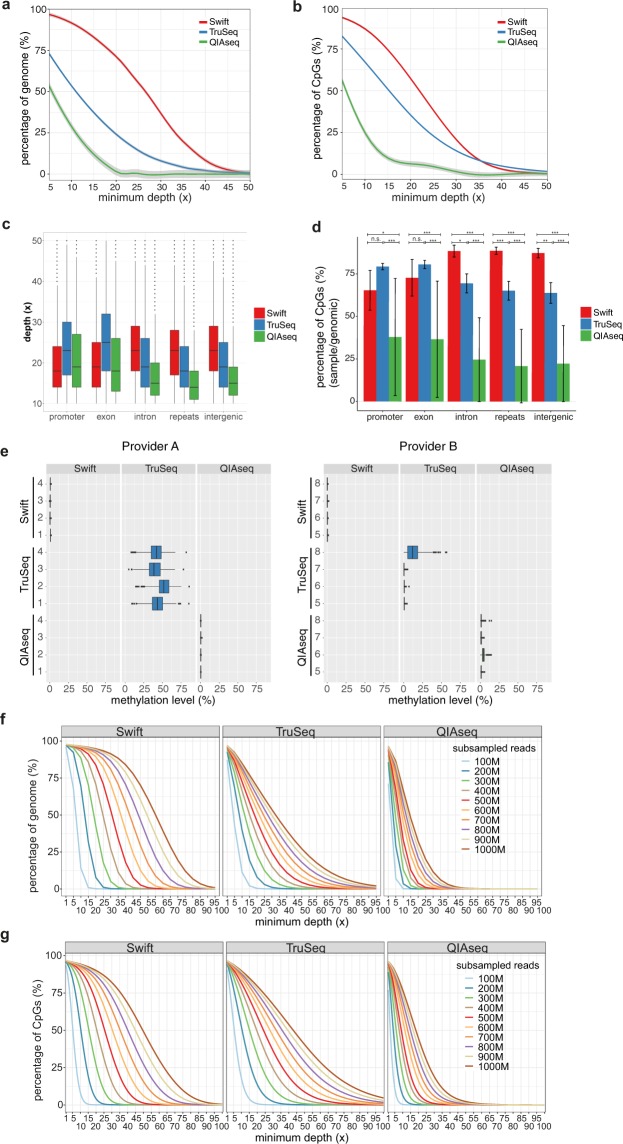
Figure 3.
Comparison of genome and CpG coverage at different minimum depths and genomic regions across library preparation methods. (a) Genome coverage at different minimum depths (5–50x). (b) CpG site coverage at different minimum depths (5–50x). For (a,b), Loess smoothing was applied, with the 95% confidence intervals indicated by gray shaded areas. (c) Distribution of depth (coverage) for CpG sites across different genomic features. Only CpGs with minimum depth between 10x and 50x (~99% of all CpG sites) were displayed in the boxplots. (d) Average percentages of CpGs covered at different genomic features. The percentages were calculated by dividing the number of CpG sites covered with a minimum depth of 10x for each genomic feature by the total number of CpG sites in the genome for the corresponding genomic feature. (e) Boxplots of methylation level for CpGs in the mitochondria across all libraries by Provider A (left) and B (right). Only CpGs with a minimum depth of 10x were included. (f) Percentage of genome covered at each minimum depth across different number of raw read pairs. (g) Percentage of CpG sites in the genome covered at each minimum depth across different number of raw read pairs. For (f,g), results obtained from downsampling analyses are shown here for minimum depths ranging from 1–100x for library sizes consisting of 100–1000 M raw read pairs. All available samples were pooled for the downsampling analyses. All libraries were sequenced on the HiSeq X platform. n.s.: not statistically significant, P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent standard error of mean.