Abstract
Myocardial infarction and ischemic stroke are the most frequent causes of death or disability worldwide. Due to their ability to dissolve blood clots, the thrombolytics are frequently used for their treatment. Improving the effectiveness of thrombolytics for clinical uses is of great interest. The knowledge of the multiple roles of the endogenous thrombolytics and the fibrinolytic system grows continuously. The effects of thrombolytics on the alteration of the nervous system and the regulation of the cell migration offer promising novel uses for treating neurodegenerative disorders or targeting cancer metastasis. However, secondary activities of thrombolytics may lead to life-threatening side-effects such as intracranial bleeding and neurotoxicity. Here we provide a structural biology perspective on various thrombolytic enzymes and their key properties: (i) effectiveness of clot lysis, (ii) affinity and specificity towards fibrin, (iii) biological half-life, (iv) mechanisms of activation/inhibition, and (v) risks of side effects. This information needs to be carefully considered while establishing protein engineering strategies aiming at the development of novel thrombolytics. Current trends and perspectives are discussed, including the screening for novel enzymes and small molecules, the enhancement of fibrin specificity by protein engineering, the suppression of interactions with native receptors, liposomal encapsulation and targeted release, the application of adjuvants, and the development of improved production systems.
Keywords: Fibrinolysis, Staphylokinase, Streptokinase, Thrombolysis, Tissue plasminogen activator, Urokinase
Abbreviations: EGF, Epidermal growth factor domain; F, Fibrin binding finger domain; K, Kringle domain; LRP1, Low-density lipoprotein receptor-related protein 1; MR, Mannose receptor; NMDAR, N-methyl-D-aspartate receptor; PAI-1, Inhibitor of tissue plasminogen activator; P, Proteolytic domain; Plg, Plasminogen; Plm, Plasmin; RAP, Receptor antagonist protein; SK, Streptokinase; SAK, Staphylokinase; t-PA, Tissue plasminogen activator
Graphical Abstract
The tertiary structure of archetypal plasminogen activator tissue plasminogen activator with colored domains and the active site shown as red sticks.
1. Introduction
1.1. Plasminogen Activators
Thrombolytics are widely used in the treatment of thrombotic diseases. The first thrombolytic drug - streptokinase - revolutionized the treatment of acute myocardial infarction [1]. Nowadays, thrombolytics are used for the treatment of acute myocardial infarction, acute ischemic stroke, pulmonary embolism, and other diseases. The advantages of thrombolytic therapy over mechanical clot-removal methods are cost-effectiveness, early onset of effect, the outlook for prehospital use, and availability in the developing countries with limited access to specialized centers providing mechanical clot-removal methods [2]. Thrombolytics activate the endogenous proenzyme plasminogen and convert it to the active form plasmin, which degrades fibrin and dissolves the blood clot (Fig. 1).
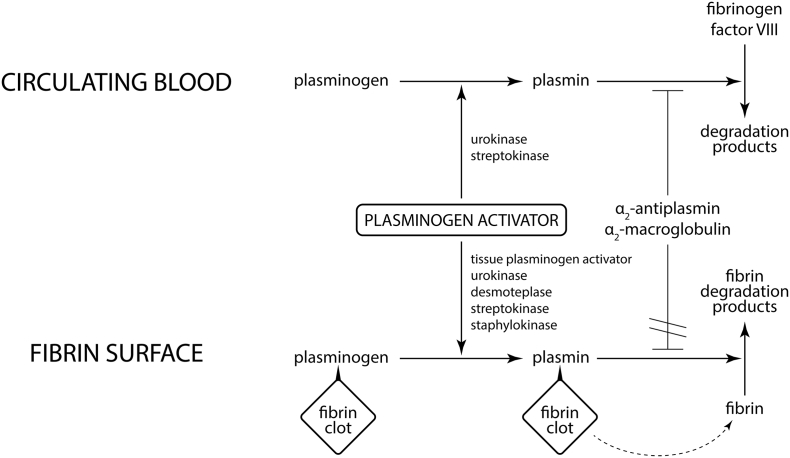
Fig. 1.
A general principle of thrombolysis. Well-characterized plasminogen activators (tissue plasminogen activator, urokinase, desmoteplase, streptokinase, staphylokinase) show a unifying principle of activating plasminogen into the active form plasmin. If plasmin is bound to the surface of a fibrin clot, it digests selectively only fibrin to form soluble fibrin degradation products. This process cannot be inhibited by α2-antiplasmin or α2-macroglobulin because the recognition site of plasmin is sterically hindered by bound fibrin. If plasmin is generated in circulating blood, it can digest fibrinogen and factor VIII instead of fibrin. This process is rapidly inhibited by α2-antiplasmin or α2-macroglobulin. Fibrinogenolysis and subsequent plasminemia caused by inhibition often lead to extensive bleeding complications. As a consequence, only plasminogen activator highly selective towards fibrin-bound plasminogen can be effective in the treatment of cardiovascular diseases.
Study of the two endogenous plasminogen activators, tissue plasminogen activator (t-PA) and urokinase, resulted in finding their numerous roles in human pathophysiology. Many side-effects were described upon administration of t-PA into the brain, mainly intracranial hemorrhage, brain edema, excitotoxicity, and neuroinflammation [[3], [4], [5]]. Consideration of these side-effects is therefore critical during both treatment and development of new therapeutics. Recent studies revealed the potential of targeting the fibrinolytic system for treating neurodegenerative and psychiatric disorders [[6], [7], [8]]. Urokinase has a crucial role in cell migration and neoangiogenesis. Specific inhibitors of urokinase have proven to inhibit metastasis of various tumors, both in vitro and in vivo [9].
1.2. Thrombolysis
Plasminogen activators can be divided by their mode of action into two groups: (i) direct and (ii) indirect. Direct plasminogen activators are eukaryotic serine proteases which activate plasminogen by its cleavage at the Arg561-Val562 bond, resulting in catalytically active plasmin. Examples of direct plasminogen activators are t-PA [10], urokinase [11] or their variants [12]. Indirect plasminogen activators are prokaryotic proteins which bind a molecule of plasminogen and induce its conformational change in a way that it can directly convert another molecule of plasminogen to plasmin. Plasmin then cleaves fibrin and eventually dissolves the thrombus. Examples of indirect plasminogen activators are streptokinase and staphylokinase.
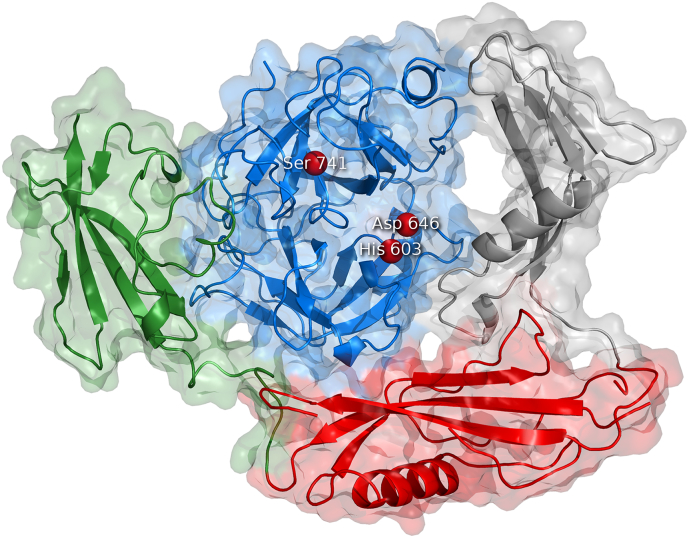
Plasminogen is a 791 amino acids long trypsin-like serine protease glycoprotein [13] with a molecular weight of 93 or 98 kDa, depending on the glycoform [14]. It can be glycosylated at Ser249, Asn289, Thr340, and Thr346 [15], and phosphorylated at Ser578 [16]. It consists of a PAN/apple domain, five kringle domains, a flexible linker where cleavage occurs, and a serine protease domain with a catalytic triad His603, Asp646, and Ser741 [15]. The kringle domains contain lysine binding sites which allow plasminogen to bind fibrin and other substrates containing N-terminal lysines. Both epsilon-aminocaproic acid and tranexamic acids bind to the lysine binding sites of plasminogen and plasminogen activators such as t-PA and urokinase. Hence, these acids act as competitive inhibitors and are used as antifibrinolytic drugs [17]. Plasminogen can be activated into plasmin by the Arg561-Val562 bond cleavage by a direct plasminogen activator [18] or via an indirect mechanism. The residues on the N-terminal side of the cleavage (1–561) form the A chain, which contains the apple domain and five kringle domains. These five domains mediate affinity to fibrin, cellular receptors and other substrates [19,20]. The A chain is linked to the B chain via disulfide bonds Cys548-Cys666 and Cys558-Cys566. The seven domains of the native form of Glu-plasminogen are in a closed activation-resistant conformation [21], which loosens up after (i) binding fibrin or (ii) removal of amino acids 1–77 by plasmin, becoming Lys-plasminogen [22]. Lys-plasminogen is less resistant to activation. The crystal structure of full-length plasminogen has been elucidated, revealing the closed conformation [23]. Full-length plasminogen promotes neoangiogenesis via extracellular matrix degradation [24]. Plasmin is cleaved by serine proteases and matrix metalloproteinases into angiostatin, a potent angiogenesis inhibitor [25]. There are two forms of angiostatin: (i) the one corresponding to domains K1–K4, and (ii) a shorter fragment of plasminogen consisting of domains K1–K3 [26]. Angiostatin inhibits angiogenesis by numerous pathways such as competing with plasmin on annexin A2/S100A10, regulating intracellular pH of endothelial cells, and interacting with other receptors on the endothelial membrane [27]. The crystal structure of the K1–K3 angiostatin has been elucidated [28].
Development of novel plasminogen activators aims to improve thrombolytic effectivity which is determined by the number of patients with successful recanalization of the clotted vessel. Thrombolytic effectivity is mediated by four main mechanisms: (i) rate of fibrin-specific activation of plasminogen by a plasminogen activator in the clot [29], (ii) penetration of plasminogen activator into the clot [[30], [31], [32]], (iii) resistance to inhibition [[33], [34], [35]], and (iv) clearance through the uptake by cell receptors [[28], [29], [30]]. Important is also time to recanalization in acute myocardial infarction [36,37] and volume of reperfused brain tissue in arterial ischemic stroke [[38], [39], [40]]. Time to recanalization is influenced by both the effectivity of the thrombolytic to degrade the blood clot and the time which elapses from the occlusion until the thrombolytic is applied [2].
Moreover, the treatment outcome is influenced by harmful side-effects: (i) bleeding complications, (ii) reocclusion [41,42], (iii) neurological side effects such as excitotoxicity [43,44] and damaging of the blood-brain barrier [45] leading to subsequent development of intracranial hemorrhage and brain edema. The side effects limit the usage of thrombolytics approved by the Food and Drug Administration in acute ischemic stroke by a therapeutic time window of 4.5 h from symptom onset [46]. Prolonging the therapeutic time window by avoiding the side-effects could lead to thrombolytic therapy for more patients. Given that time to therapy is a crucial predictor of the outcome, striving for less pronounced side-effects or pre-hospital therapy could lead to earlier application of thrombolytics, and therefore a better outcome [2]. Immunogenicity is another major concern for non-human plasminogen activators.
The most commonly clinically used thrombolytics is t-PA [12]. One of the advantages of t-PA (Table 1) over the first-generation thrombolytics is its fibrin specificity [47]. The fibrin specificity is directly connected to organization of the domains within protein molecules (Fig. 2). Non-specific thrombolytics, e.g., β-hemolytic streptococcal streptokinase and the second endogenous thrombolytic, urokinase, cause plasma fibrinogen depletion and bleeding complications (Fig. 3). The disadvantage of high fibrin specificity of t-PA is front-like lysis, where most of the t-PA binds to the first micrometers of the fibrin clot. This binding hinders the penetration of t-PA inside and disables the lysis in the whole volume of the clot, causing non-optimal effectivity. This uneven lysis pattern can lead to the formation of smaller clots which are released from the blood vessel wall and can cause reocclusion [30,32,48]. The third generation of thrombolytics are: (i) derivatives of t-PA engineered for enhanced half-life, penetration into a clot, fibrin specificity and resistance to inhibition, e.g., reteplase, tenecteplase, duteplase, monteplase, lanoteplase, pamiteplase, amediplase [12], (ii) desmoteplase from the vampire bat saliva, and (iii) fibrin-specific prokaryotic staphylokinase.
Table 1.
A comparison of three generations of thrombolytics in terms of their structure and biological properties.
| Thrombolytics | Domains | Protein data bank IDs | Mode of action | Fibrin affinity | Fibrin selectivity | Half-life [min] | Risk of inhibition | Risk of side-effects |
|---|---|---|---|---|---|---|---|---|
| First generation | ||||||||
| Streptokinase | α, β, γ | 1BML | Indirect | None | None | 10 | Low | High |
| Urokinase | EGF, K1, P | 4DVA, 2I9B | Direct | None | Low/nonea | 8 | High | High |
| Second generation | ||||||||
| Tissue plasminogen activator | F, EGF, K1, K2, P | 1TPG, 1TPK, 1BDA | Direct | Moderate | Moderate | 4.5 | High | Low |
| Third generation | ||||||||
| Desmoteplase | F, EGF, K1, P | 1A5I | Direct | High | High | 138 | Low | Low |
| Staphylokinase | α | 1BUI | Indirect | None | High | 6 | Low | Moderate |
Prourokinase is fibrin-selective, urokinase is not fibrin-selective.
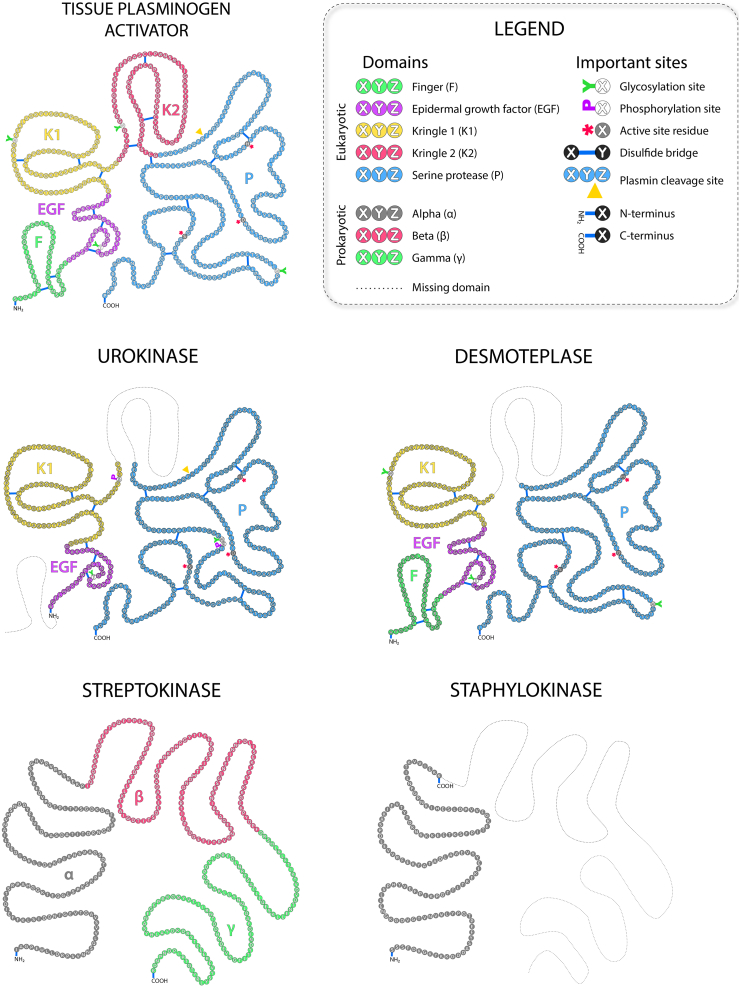
Fig. 2.
Comparison of domain organization in eukaryotic and prokaryotic plasminogen activators. The corresponding domains for the eukaryotic proteins t-PA, urokinase, and desmoteplase – finger F, epidermal growth factor EGF, kringle K1, kringle K2, serine protease P – are depicted with individual colors. Glycosylation sites are marked with a green Y symbol, phosphorylation sites with a purple P symbol, catalytic residues with a red asterisk *, disulfide bridges with a blue line, and the site of the proteolytic cleavage is marked with a yellow triangle Δ. The individual domains α, β, and γ are marked with corresponding colors for prokaryotic proteins streptokinase and staphylokinase. The missing domains in homologous proteins are illustrated with a dashed line.
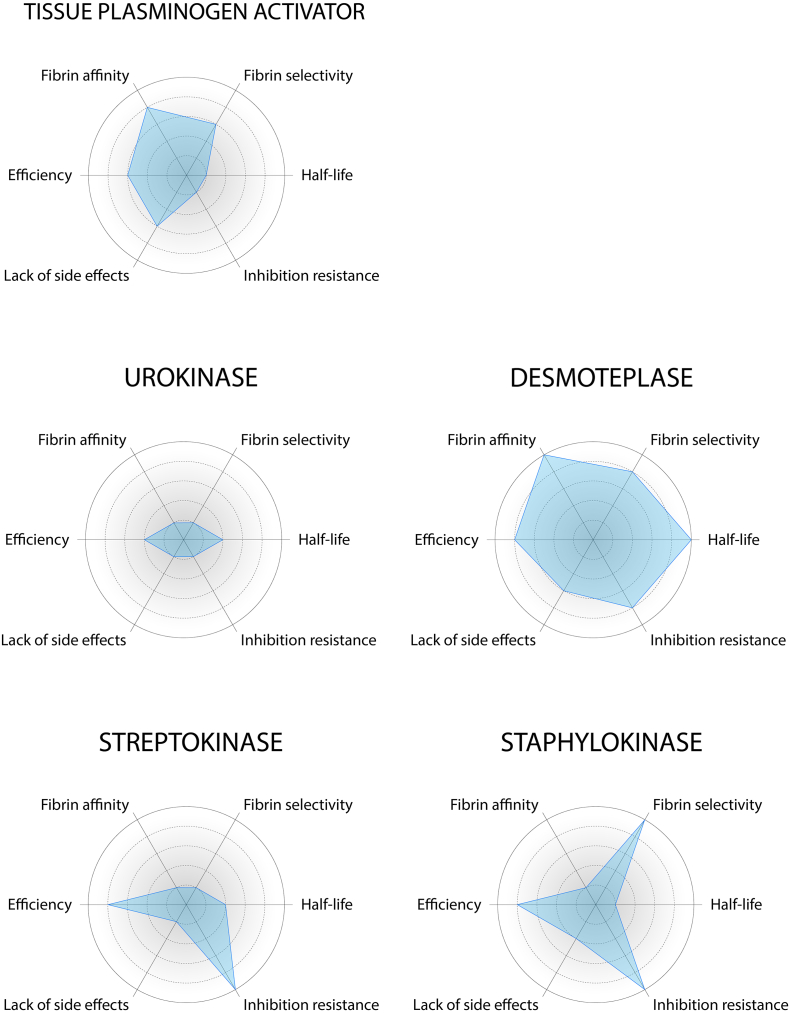
Fig. 3.
Comparison of key properties of eukaryotic and prokaryotic plasminogen activators. The size of the blue area quantifies the overall potential for clinical use of the protein (large area = high potential). The radial plots are based on the values published in the scientific articles cited throughout this review article.
2. Tissue Plasminogen Activator
2.1. Biological Function
Besides fibrin-specific thrombolysis, t-PA also has many roles in the brain (Fig. 4). A high concentration of t-PA in the bloodstream during therapeutic thrombolysis can result in deleterious side-effects. t-PA is a modulator of cerebral blood flow and blood-brain barrier permeability in response to neuronal activity and facilitates memory formation and response to brain injury [3,29,45,[49], [50], [51], [52], [53]]. Both t-PA and the t-PA-PAI-1 complex have effects on blood vessel tone. The t-PA-PAI-1 complex induces vasodilation whereas t-PA alone induces vasoconstriction. Vasodilation can have beneficial effects of better penetration into the blood clot, which could be a downside of tenecteplase [54].
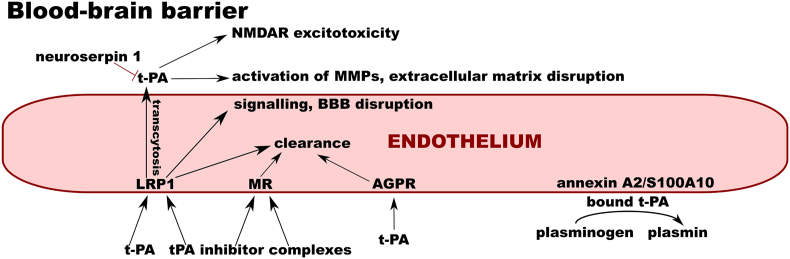
Fig. 4.
Interactions of t-PA on the blood-brain barrier. On the endothelial cell surface, tissue plasminogen activator (t-PA) binds annexin A2/S100A10. This leads to plasminogen activation on the cell surface. The free form, as well as inhibitor complexes of t-PA, are endocytosed by asialoglycoprotein receptor (AGPR), mannose receptor (MR), and low-density lipoprotein receptor-related protein 1 (LRP1), leading to their clearance. T-PA can cross the intact blood-brain barrier (BBB) via LRP1-dependent transcytosis. In the brain parenchyma, it can activate matrix metalloproteinases and interact with the N-methyl-D-aspartate receptor (NMDAR). These effects are inhibited by neuroserpin 1, the inhibitor of tPA in the brain parenchyma.
2.2. Molecular Structure
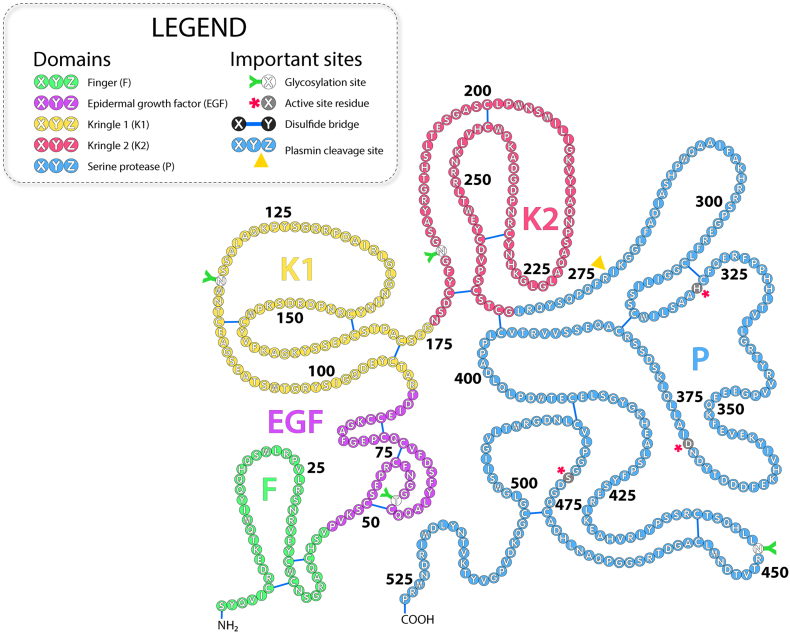
T-PA is a 527 amino acids long glycoprotein belonging to the trypsin-like serine protease family (Fig. 5). It exists in two glycoforms: type 1 and type 2 t-PA with a molecular weight of 66 or 63 kDa, respectively. Type 1 has oligosaccharides bound at both Asn448 and Asn184, while type 2 is glycosylated at Asn448 only [55]. T-PA carries 17 disulfide bridges which make it hard to express in prokaryotic systems and many studies are focused on improvement of its production. T-PA is composed of five domains [56]: (i) the fibronectin-like finger F domain (residues 1–49), (ii) the epidermal growth factor EGF domain (50–87), (iii) kringle K1 domain (88–175), iv) kringle K2 domain (176–256), and v) the serine protease P domain (257–527, Fig. 6).
Fig. 5.
The primary structure of tissue plasminogen activator. Individual domains – finger (F), epidermal growth factor (EGF), kringle 1 (K1), kringle 2 (K2), serine protease (P) , important residues, cysteine-bridges, cleavage sites and sites of post-translational modification are highlighted as stated in the legend.
Fig. 6.
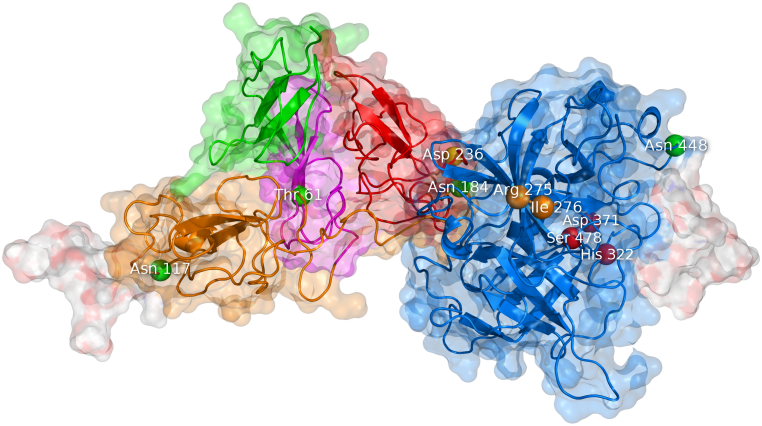
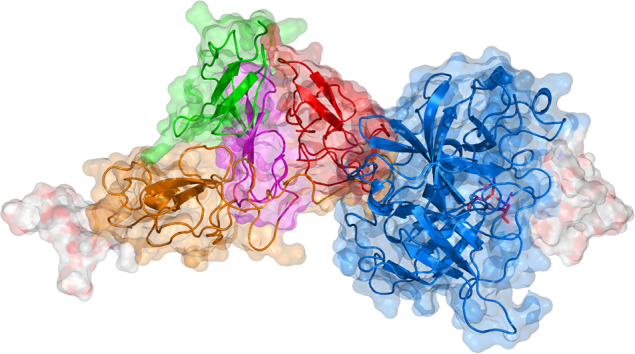
Theoretical model of the tertiary structure of tissue plasminogen activator. The visualization is based on the model kindly provided by Ashish and coworkers [78]. T-PA is composed of five domains: finger F (green), epidermal growth factor EGF (purple), kringle K1 (orange), kringle K2 (red), and protease P (blue). The molecule contains four glycosylation sites labeled as green beads: Thr61, Asn117, Asn184, and Asn448. The molecule also has complex oligosaccharides attached to Asn117 and Asn448, which are shown as white surface. The K2 domain contains a lysine binding site at Asp236 (yellow bead). The P domain can be cleaved into two chains in between Arg275 and Ile276 (orange beads). The catalytic triad of the P domain is His322, Asp371, and Ser478 (red beads).
The domain F contributes to about 80 % of the affinity to fibrin [57]. It also binds to annexin A2/S100A10 located on the cell surface [58] and is involved in the low-density lipoprotein receptor-related protein 1 (LRP1) binding together with the domains EGF and K1 [59]. The EGF domain activates the EGF receptor [60] and is fucosylated on Thr61 [61]. The domain K1 binds to the mannose receptor (MR) expressed in liver endothelial and Kupffer cells by high-mannose oligosaccharide linked to Asn117 [62]. The domain K2 plays a crucial role in the stimulation of proteolytic activity by fibrin [63] and by other lysine-containing substrates [64]. The domain K2 contains a negatively charged lysine binding site around Asp236 [65] which binds fibrin, amyloid beta aggregates [66], N-methyl-D-aspartate receptor (NMDAR) [67], and others via nonspecific interaction with C-terminal and intra-chain lysines [68]. The domain P is catalytically active and carries a catalytic triad of His322, Asp371, and Ser478 [69].
2.3. Mechanism of Activation and Specificity
Most serine proteases are expressed in an inactive, single-chain form and must be cleaved to the two-chain active form. T-PA is cleaved between Arg275-Ile276 by plasmin [70] into N-terminal “A chain” and the C-terminal “B-chain” which are connected by a single disulfide bond between Cys264-Cys395 [71]. While other serine proteases are more than 107 times less active in the single-chain form, t-PA has low zymogenicity with only 5–10-fold increase in activity upon cleavage [72]. The catalytic activity of t-PA is regulated by a dynamic conformational equilibrium of the activation domain [39,73,74] restricting the binding of plasminogen in the inactive states. Activation domain contains the catalytic triad, an oxyanion hole and an S1 specificity pocket [69]. Active conformations are favored when a salt bridge is formed between Asp477 and the N-terminal of the B-chain of two-chain t-PA. Low zymogenicity of single-chain t-PA is caused by substitute salt bridge of Lys429 and Asp477, which leads to an active conformation.
T-PA has a general trypsin-like specificity, preferentially cleaving peptide bonds after Arg and a small or a hydrophobic residue. The plasminogen cleavage site is a Cys-Pro-Gly-Arg-Val-Val-Gly-Gly-Cys cyclised by the flanking cysteines. The active site cleft of t-PA is specifically well suited for cleaving it since linear peptides mimicking the plasminogen cleavage site are cleaved 104 times less efficiently than plasminogen [75]. The adaptation to the cyclic sequence in plasminogen is determined by loops around the active site [72] and the specificity for plasminogen is enhanced by a hydrophobic exosite at the residues 420–423 [76].
2.4. Binding and Stimulation by Fibrin
T-PA variants with: (i) uncleavable single-chain form [67,77], (ii) restored zymogen triad [77], or (iii) the single-chain form salt bridge disrupted [70,72], are stimulated by fibrin to a similar activity as the wild type t-PA. The property of fibrin stimulation limits systemic fibrinogen depletion. All five domains of t-PA account for fibrin binding, with the strongest effect of the F domain followed by the K2 domain. The stimulation of activity is mediated by the heavy-chain domains via co-localization of t-PA and plasminogen on fibrin in a productive orientation and by increasing the catalytic rate of t-PA.
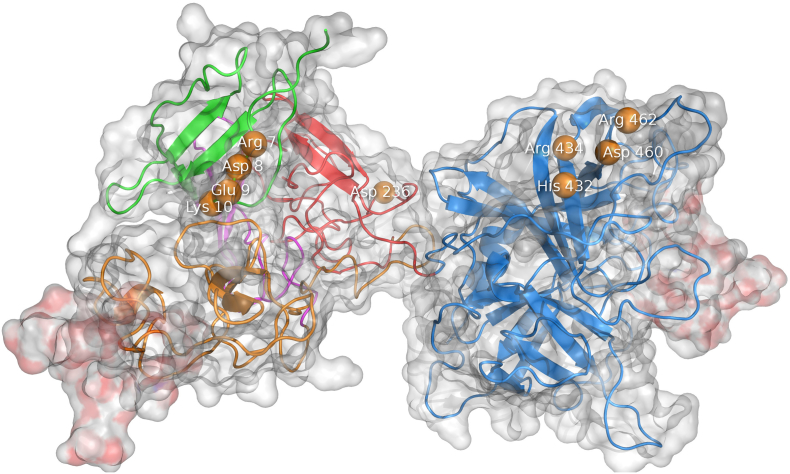
The glycosylation at Asn184 insulates contacts between the F, K2, and P domains, in the less active type 1 [78]. The orientation of the F, E, K2, and P domains, towards each other is mediated by the domain K1, which explains the deleterious effect of deletion of any domain on activity and stimulation by fibrin [79]. Binding of the penta-L-lysine peptide between the K2 and P domains stimulated the activity by increasing the number of inter-domain interactions and reducing protein dynamics [78]. Domain deletion studies have reported inconsistent results, some indicating the K1 domain is not significantly involved in binding [57,80], whereas others suggested that one kringle domain, be it K1 or K2, can facilitate fibrin binding [63]. The F domain contains patch of charged residues Arg7, Asp8, Glu9 and Lys10, while the P domain (Fig. 7) contains a fibrin binding patch of the residues His432, Arg434, Asp460, and Arg462 [69,81].
Fig. 7.
The fibrin binding sites of tissue plasminogen activator. The residues interacting with fibrin are shown as orange beads. Two charged patches are involved in binding of fibrin: (i) Arg7–Lys10 in the t-PA’s finger domain and (ii) residues His432, Arg434, Asp460, and Arg462 in the serine protease domain. The lysine binding site at Asp236 binds both C-terminal and intra-chain lysines of partially degraded fibrin.
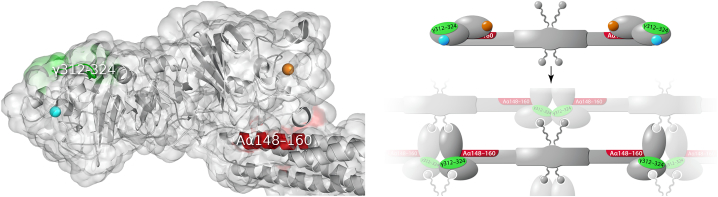
T-PA’s high selectivity for fibrin over fibrinogen [82] is mediated by cryptic sites present in fibrin [83]. Cross-linked αC domain fragments Aα392–610 contain high-affinity binding sites for t-PA and plasminogen [84]. Their binding is lysine-dependent which suggests the domain K2 binds these sites. A low-affinity binding site on fibrin (γ312–324, Fig. 8) is positioned next to the A knob binding cavity [85]. When A knob binds, the β globular domain is pulled away and the buried binding site which can bind of t-PA or plasminogen (Aα148–160) is exposed [86,87]. Similarly, binding of B knobs also makes the Aα148–160 site accessible whereas synthetic B knobs showed fibrinolysis inhibition [88]. Limited digestion by plasmin reveals new C-terminal lysines which bind t-PA and plasminogen, resulting in stimulation of fibrinolysis [89,90].
Fig. 8.
Tertiary and quaternary structure of fibrin. (Left panel) The C-terminal part of fibrinogen (PDB ID 3GHG). The α chain of fibrinogen contains a site which can bind either t-PA or plasminogen at the residues Aα148–160 (red). Residues γ312–324 form a site which binds t-PA (green). The sites are situated very close to the A-hole (blue bead) and B-hole (orange bead) which in fibrin polymerization bind the A- and B-knobs, respectively. (Right panel) The t-PA and plasminogen binding sites, and the A- and B-hole are highlighted as in panel A. Upon polymerization, the A knobs interact with the A holes, the B knobs with the B holes, and the globular C-terminal parts of the α and β chain get spatially reoriented, exposing the buried Aα148-160 site.
2.5. Inhibition
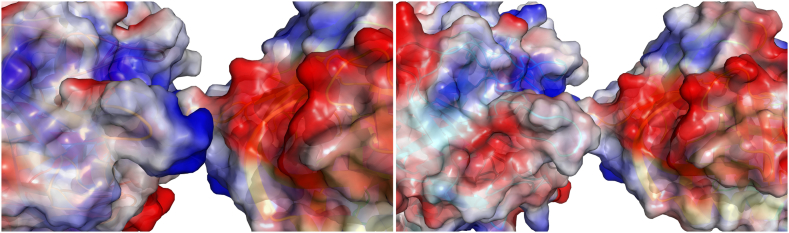
T-PA is irreversibly inhibited by proteins of the serpin family: PAI-1, PAI-2, protease-nexin-1, and neuroserpin [[91], [92], [93], [94]]. These inhibitors bind by their reaction center loop into the active site of t-PA [94]. T-PA cleaves the bond of its major inhibitor PAI-1 and remains covalently bound by the catalytic serine. The interaction with PAI-1 (Fig. 9) is determined by the positively charged loop 296–302 and the residue Arg304 [95,96]. The 296–302 tetra-alanine substitution in tenecteplase [97] or charge-reversing mutations in this region [98] can substantially decrease the inhibition. Other strongly interacting residues representing inhibition hot-spots are Gln325, Glu326, Asp365, and Tyr368. Especially, Tyr368 protrudes to the active site cleft of t-PA and restricts the S2 specificity pocket which causes resistance to most canonical serine protease inhibitors [69]. Tyr368Leu mutation improves the resistance of t-PA to PAI-1 without compromising the activity with plasminogen [99]. Mutation Ala419Tyr introduced into the hydrophobic pocket of t-PA formed by Ala419, Gln475 and Gly501 [96] increased PAI-1 resistance 30-fold and plasminogen activation 5-fold [100].
Fig. 9.
Interactions of t-PA and tenecteplase with the plasminogen activator inhibitor-1. (Left panel) The interface between t-PA (left) and the plasminogen activator inhibitor-1 (right) with their surface colored according to the surface charge (PDB ID 5BRR). T-PA contains a positively charged loop of residues 296–302 which fits into a negatively charged cleft on PAI-1. (Right panel) The catalytic domain of tenecteplase (left) carries the 296–299 tetra-alanine substitution which weakens its interaction with PAI-1 (right).
2.6. Interaction with Receptors and Clearance
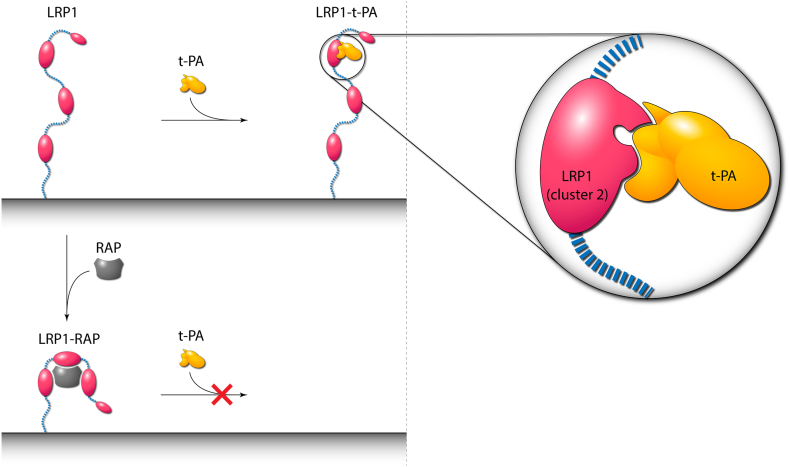
Interactions of t-PA with receptors mediate many biological functions (Fig. 10). The most important is binding to liver cell receptors (LRP1), which are responsible for the short half-life of only 4.5 min [101]. LRP1 plays a major role in clearance, followed by MR [62], galectin [102], and potentially by the asialoglycoprotein receptor [55]. Gp330 is highly homologous to LRP1 and could be responsible for the clearance of t-PA in the kidney [103,104]. The binding sites for both free t-PA and the t-PA-PAI-1 complex on LRP1 are on complement repeats of cluster 2 of LRP1 [105]. Binding can be inhibited by the receptor antagonist protein (RAP), which inhibits the binding both by competing for the part of the binding site and allosterically [103,104,106]. The ligand-binding mechanism of LRP1 employs aspartates around Ca2+ ions [101,[107], [108], [109], [110]] which bind exposed lysines. In t-PA and desmoteplase [111], the LRP1 binding sites could be present in the F, E, and K1 domains [58,59,112]. Tyr67Gln mutation impairs binding via an extra oligosaccharide inserted, which hinders the interaction with LRP1 [113]. The effects of LRP1 interaction with t-PA mediate pleiotropic roles of t-PA in neurophysiology [29,49], but also contribute to neurological side-effects of therapeutically used t-PA.
Fig. 10.
Schematic representation of binding of t-PA to low-density lipoprotein receptor-related protein 1 (LRP1) and binding of receptor-associated protein (RAP). (Left panel) Low-density lipoprotein receptor-related protein 1 (LRP1) binds to t-PA via cluster 2 (red oval) of its structure. In contrast, receptor-associated protein (RAP) possesses three binding domains, binds three clusters of LRP1 simultaneously and induces conformational changes that prevent the binding of t-PA. RAP does not compete for the same binding site but allosterically makes the site inaccessible for t-PA binding. (Right panel) A close-up view of the LRP1-t-PA complex. The binding of t-PA to LRP1 is enhanced by an avidity effect of two separate binding sites of t-PA that interact with distinct complement repeats of cluster 2 of LRP1.
All these effects on the nervous system are more pronounced when t-PA is used in the treatment of acute ischemic stroke. The MR is proven to bind the high-mannose oligosaccharide attached to Asn117. Deletion of K2 reduces NMDAR-mediated neurotoxicity [114] as well as mutations in the positively charged loop (296–299) [115]. On the other hand, proteolytic activity is not necessary for NMDAR activation so the specific interaction remains unclear [44,115]. Desmoteplase does not contain a lysine binding site and is not neurotoxic [116]. Interestingly, only the single-chain form of t-PA activates NMDAR [43]. Since t-PA interacts with Annexin A2/S100A10 via C-terminal lysines, the lysine binding site on K2 domain is likely the determinant of the binding. Annexin A2/S100A10 also binds plasminogen at the site of the cytoplasmic membrane and is a source of plasminogen activation on the cell surface [117]. Binding to endothelial cells by t-PA’s kringle domains has been shown to inhibit neoangiogenesis [[118], [119], [120]]. The binding of t-PA to the epidermal growth factor receptor protects neurons in ischemic conditions [60]. Binding to annexin A2/S100A10 tetramer on the surface of vascular endothelial cells enhances the activation of plasminogen on the cell surface and activates nuclear factor κB involved in immune response [[121], [122], [123]]. Both of these effects could cause problems in thrombolytic therapy: (i) reocclusion complications and (ii) inflammation.
2.7. Protein Engineering
One aim of protein engineering of t-PA was to increase its half-life. A deletion mutant reteplase retains only the K2 and P domains [124] and has the tetra-alanine substitution. Half-life is thus enhanced by the elimination of binding to the MR, decreased binding to LRP1 and resistance to PAI-1. Tenecteplase also contains the tetra-alanine in addition to Thr103Asn which introduces a complex oligosaccharide, causing steric hindrance of clearance receptors, and Asn117Gln eliminating MR recognition [125]. The tetra-alanine also increases fibrin specificity [126]. Monteplase with Cys84Ser in the EGF domain has a half-life of 23 min [128]. Duteplase is a Met245Val mutant produced exclusively as a two-chain form and possesses slower clearance rate than single chain t-PA [127]. Lanoteplase has a prolonged half-life to 37–45 min [128] due to the absence of a large part of the F domain, whole EGF domain and the Asn117Gln substitution [129]. Pamiteplase is a K1 domain deletion mutant with a half-life of 30–47 min. It also contains the Arg275Glu mutation which renders it uncleavable to the two-chain form [130,131]. Amediplase is a chimeric protein composed of the first 3 residues of the F domain and the K2 domain of t-PA, combined with residues 159–411 of the P domain of urokinase. The lower fibrin affinity enables better penetration into the clot. It has a half-life of 30 min [12], likely because of the absence of F, EGF, and K1 domains. A variant consisting of the GHRP peptide mimicking the B knob of fibrin, K2, and P domain has an increased half-life as well as improved binding to fibrin compared to reteplase [128]. Arg275Ser mutation is preventing the conversion to the two-chain form and thus NMDAR-mediated neurotoxicity. The variant also has the lysine binding site in K2 defunct via the Trp254Arg mutation [114]. Higher fibrin binding and fibrin stimulation were achieved by substituting the finger domain of t-PA by that of desmoteplase and deleting the K2 domain [132].
3. Urokinase
3.1. Biological Function
Urokinase accelerates thrombolysis initiated by t-PA and was tested as a synergistic therapy [48,[133], [134], [135], [136]]. The implications of urokinase in cancer make it a target for the development of urokinase-specific inhibitors [137] or disrupting interactions with its receptors [138,139]. Urokinase is primarily associated with plasminogen activation on the cell surface and is involved in neoangiogenesis [140], degradation of the extracellular matrix by plasmin [141], and activation of matrix metalloproteinases [142,143]. These mechanisms confer the roles of urokinase in cell migration [144], tumor metastasis [145], cell adhesion [146], and proliferation of various cancer types [147,148].
3.2. Molecular Structure
Urokinase is a 411 amino acids long protein with a molecular weight of 54 kDa [71], having a 40 % identity with t-PA [149]. It consists of the EGF domain (1–49), a K domain (50–131) lacking a lysine binding site [150], a flexible linker (132–158), and a serine protease domain P (159–411) with the catalytic triad of His204, Asp255, and Ser356 (Fig. 11). Phosphorylations on Ser138 and Ser303 contribute to the signaling through the urokinase receptor [151]. Urokinase is fucosylated on Thr18 [152] and N-glycosylated on Asn304 [153]. The enzyme is secreted as prourokinase, which is cleaved by plasmin and other proteases at Lys158-Ile159 to a two-chain form called high molecular weight urokinase [154]. Additional cleavage at Lys135-Lys136 generates the N-terminal EGF-K fragment and low molecular weight urokinase.
Fig. 11.
The catalytic serine protease domain and epidermal growth factor domains of urokinase complexed with the urokinase receptor. (Left panel) The serine protease domain of urokinase (PDB ID 4DVA) with its triad of His204, Asp255, and Ser356 shown as red beads. (Right panel) The EGF domain of urokinase (PDB ID 2I9B) is shown as violet surface and the urokinase receptor as cyan surface. The omega loop of the amino terminal fragment which binds the urokinase receptor is highlighted in red.
3.3. Mechanism of Activation and Specificity
The conversion to two-chain form increases inter-domain flexibility [155] which enables to interact more efficiently with its substrates and improves the activity of urokinase about 100 times [156]. Cofactors shift the equilibrium towards the formation of an active catalytic site [157] or could improve the interaction of the new Ile159 N-terminus with Asp370, analogically to the activation of t-PA [158,159].
3.4. Binding and Stimulation by Fibrin
Prourokinase cleaves relaxed plasminogen bound to fibrin [160]. Interestingly, prourokinase is stimulated only by fibrin fragment E, but not fragment D, which could be due to different conformations adapted by plasminogen when bound there. Mutation of Pro309 can make urokinase stimulated by both fragments E and DD [161]. Two-chain form urokinase is not fibrin-selective and causes systemic fibrinogen depletion.
3.5. Inhibition
Urokinase is inhibited by PAI-1, PAI-2, protease-nexin [162], and thrombin [163]. As in t-PA, the positively charged loop around His200 contributes significantly to the recognition by serpins [164]. The aim of understanding the specific inhibition of urokinase has led to crystallographic and inhibitor screening studies [[164], [165], [166], [167]].
3.6. Interaction with Receptors and Clearance
Urokinase’s half-life of 8 min in the human bloodstream is mediated by the receptor LRP1 and asialoglycoprotein [168]. LRP1 internalizes either free urokinase or the trimeric complex with PAI-1/PAI-2 and the urokinase receptor. The binding to LRP1 is probably mediated by domains EGF and K [[169], [170], [171], [172]]. Plasminogen activation on the cell surface is mediated by Ω loop of the EGF domain of urokinase binding to the urokinase receptor [[173], [174], [175], [176]]. When bound to the urokinase receptor, urokinase regulates cell adhesion by cleavage of vitronectin [146]. The modulation of chemotaxis via interaction with integrins is probably related to the presence of the flexible linker [177,178].
3.7. Protein Engineering
Chimeric combinations of t-PA and urokinase domains possess poor fibrin affinity [179]. Amediplase has poor fibrin affinity due to the interaction of the P domain from urokinase with both K domains from t-PA. On the other hand, it has better penetration into the clot and a longer half-life [12]. The mutation Lys300His has 10 times lower amidolytic activity in the single-chain form and 2 times higher in the two-chain form [180,181].
4. Streptokinase
4.1. Biological Function
Streptokinase is an exocellular plasminogen activator of the prokaryotic origin first described in 1933 [182]. It is naturally produced by the strains of β-hemolytic streptococci which utilize it for overcoming the host’s defensive fibrin barrier and for promoting bacterial metastasis and colonization [183,184]. There are several streptokinases from different streptococci which vary in their structure, but the only variant of streptokinase currently used as a thrombolytic agent originates from the group C streptococci (streptokinase SK-H46A from Streptococcus equisimilis strain H46A) and lacks a significant stimulation by fibrin [185,186]. Despite its moderate half-life in vivo (approximately 10 min) and significantly lower cost, the largest disadvantages are the lack of fibrin specificity and immunogenicity due to its bacterial origin [47,[187], [188], [189]].
4.2. Molecular Structure
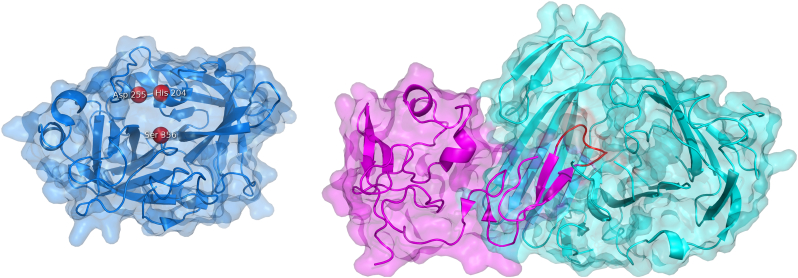
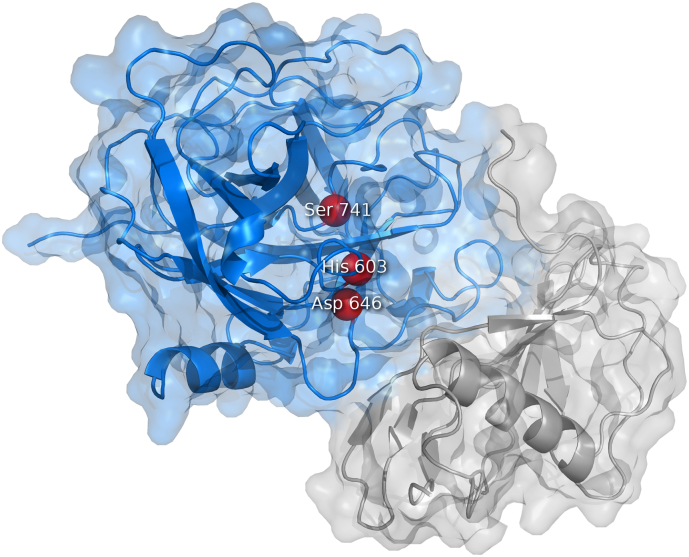
The molecule of streptokinase consists of 414 amino acids and the biological unit forms the monomeric protein with the molecular weight of 47 kDa [190,191]. The isoelectric point is 4.7, while the pH optimum is between 7.3 and 7.6 [187,191]. Nuclear magnetic resonance and circular dichroism studies [152,192] alongside with the crystallographic analysis of the microplasmin-streptokinase complex [193] showed that streptokinase contains three β-grasp domains – α, β, and γ (Fig. 12). The domain α (residues 1–150) mainly mediates the formation of the plasminogen active conformation within the streptokinase-plasminogen complex [[194], [195], [196], [197], [198], [199], [200]] and is important for the substrate recognition [194,197,198,[201], [202], [203]]. The domain β (residues 151–287) is responsible for high-affinity binding during the activation of the plasminogen “partner” [197,[204], [205], [206]] but it also facilitates the plasminogen “substrate” binding and processing [[207], [208], [209], [210], [211]]. Finally, the domain γ (residues 288–414) is involved in stabilizing the streptokinase-plasminogen complex and in inducing its proteolytic activity [193,197,201,204,208,[212], [213], [214]]. Although having one or two major functions, each domain participates in all the steps of plasminogen activation due to the high level of cooperativity [206,215,216].
Fig. 12.
The binary complex of streptokinase with bound plasmin. The complex of streptokinase and the catalytic domain of plasmin (blue) (PDB ID 1BML). Streptokinase contains three β-grasp domains embracing the plasmin catalytic domain: domain α (gray), domain β (red), and domain γ (green). The catalytic triad of plasmin His603, Asp646, and Ser741 are shown as red beads.
4.3. Mechanism of Activation and Specificity
Compared to human-derived plasminogen activators, streptokinase represents a thrombolytic agent with an indirect mechanism of plasminogen activation (Fig. 13), i.e., it does not possess enzymatic activity. The activation process occurs in three fundamental steps including (i) formation of an equimolar (1:1) complex with the molecule of plasminogen (“partner molecule”), (ii) induced intramolecular rearrangement of the complex generating an enzymatically active structure, and (iii) amidolytic conversion of other free plasminogen molecules (“substrate molecules”) to plasmin by the active streptokinase-plasminogen complex [201,[217], [218], [219]].
Fig. 13.
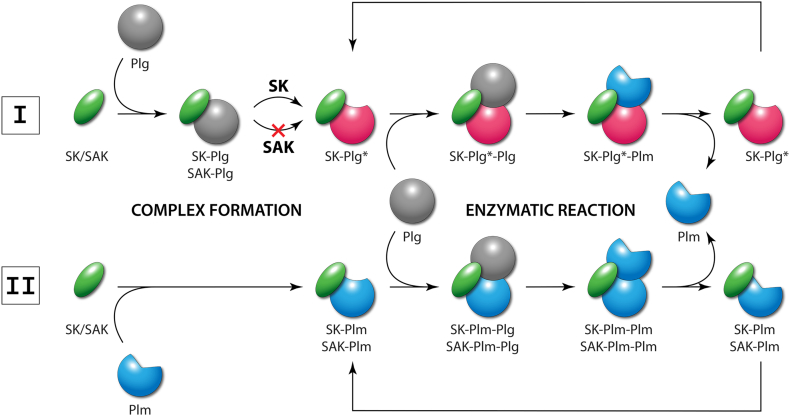
Schematic representation of the indirect mechanism of plasminogen activation. Pathway I: Streptokinase (SK) or staphylokinase (SAK) form an equimolar complex with free plasminogen (Plg) but only SK is able to form an enzymatically active complex (SK-Plg*) with an open conformation of Plg. The activated complex then catalyzes the conversion of another free Plg molecule to active plasmin (Plm). Pathway II: An equimolar complex of SK/SAK with Plm is formed with higher affinity, does not require conformational activation and is enzymatically active with either SK or SAK. This complex (SK-Plm/SAK-Plm) catalyzes the same conversion of Plg to Plm as SK-Plg*. Compared to SK, SAK can activate Plg only via Pathway II.
The detailed mechanism of all these steps was intensively studied and the current understanding assumes the complex behavior of all the participating structures as follows:
-
1.
The β domain of streptokinase interacts with the lysine binding site of the kringle 5 domain of the “partner” plasminogen molecule which, originally closed, becomes more extended and allows all the three domains of streptokinase to associate with the catalytic domain of plasminogen [197,206,220,221]. Affinity towards plasminogen during the initial step is further enhanced by the C-terminal Lys414 which interacts with the plasminogen’s kringle 4 domain [214,222,223].
-
2.
Within the formed encounter complex, the N-terminal amino group of Ile1 residue of streptokinase forms a salt bridge with Asp740 of plasminogen [195,199,200,207] and simultaneously, additional salt bridges between the γ domain of streptokinase and the catalytic domain of plasminogen are created [201,212]. These interactions cause conformational changes, open the active site of plasminogen and make the overall complex enzymatically active, forming so-called “virgin enzyme”, marked as SK-Plg* in Fig. 13 [197,201,217,218].
-
3.
The conformation of the active complex allows binding of another molecule of plasminogen (“substrate”) resulting in a transient ternary intermediate [201,211]. The “substrate” molecule is bound predominantly by the α domain [197,[201], [202], [203]] while the 250-loop of the streptokinase’s β domain (residues Arg253, Lys256, and Lys257) interacts with the kringle 5 domain of the plasminogen substrate via its lysine binding site [207,208,210,211,220].
-
4.
Finally, the Arg561-Val562 peptide bond of the “substrate” molecule is hydrolytically cleaved by the “partner’s” catalytic triad His603, Asp646, and Ser741, and the final molecule of active plasmin is formed and released [201,217,219]. Regions 88–97, 164–186, and 314–342 of streptokinase were reported to be important for the plasminogen substrate processing during this final step of activation [208,209,213,215,224].
-
5.
When traces of plasmin molecules are generated, streptokinase tends to form the streptokinase-plasmin complex preferentially due to the approximately three orders of magnitude higher affinity towards plasmin compared to plasminogen. This complex does not require any conformational changes for activation as in the case of streptokinase-plasminogen but only narrows the broad substrate specificity of the plasmin partner towards the plasminogen substrate [199,219,221,225].
4.4. Binding and Stimulation by Fibrin
Due to the α domain, streptokinase does not require fibrin and activates plasminogen independently [186,194,200]. Stimulating experiments confirmed that neither fibrin nor fibrinogen had stimulatory effects on activation, preventing targeted thrombolysis on the surface of fibrin clots [185,186]. The activation independent of fibrin can lead to bleeding complications which represents one of the main drawbacks of the currently used streptokinase variant.
4.5. Inhibition
Unlike t-PA and urokinase, streptokinase has no natural inhibitor present in a human body. Since it is not homologous to eukaryotic plasminogen activators, it is not recognized by traditional plasminogen activator inhibitors [47]. Similarly, the streptokinase-plasminogen/streptokinase-plasmin complex is not inhibited by α2-antiplasmin [[226], [227], [228]]. However, streptokinase’s non-human origin causes the production of neutralizing antibodies by the human immune system when used as a drug. Thus, repeated administration decreases streptokinase’s thrombolytic efficiency and can lead to a serious allergic response [188,189].
4.6. Protein Engineering
Different approaches were tested to enhance fibrin specificity of streptokinase. Deletion of the α domain prevented streptokinase to interact with the closed conformation of plasminogen. Instead, it formed the streptokinase-plasmin complex exclusively and was able to activate only the extended conformation of plasminogen bound to the fibrin surface. This step made streptokinase an activator resembling the molecule of staphylokinase [186,194,200,229]. An alternative strategy was based on using fibrin-specific streptokinases from different Streptococcus strains or on shuffling activators’ domains to combine their properties. While keeping the original activity, the fibrin-specificity of these new molecules was comparable or even higher than for t-PA [185,[230], [231], [232]].
Different engineering attempts were focused on increasing the half-life of streptokinase. Strategies were based on finding that during the process of plasminogen activation, the generated plasmin non-physiologically cleaves streptokinase into three polypeptide chains (residues 1–59, 60–386, and 387–414) leading to drop in activity [233,234]. Preventing the cleavage by mutagenesis at the identified positions (Lys59Gln/Glu, Lys386Gln) resulted in a 21-fold increased half-life without affecting activity [[235], [236], [237]]. Alternative strategies based on combinatorial mutagenesis [14,238], PEGylation [[239], [240], [241], [242]], glycosylation [235,243], or lipidification [244] yielded streptokinase variants with improved thrombolytic activity, decreased immunogenicity, and higher half-life.
A very successful variant of streptokinase is its acylated complexed form known as anistreplase. Anistreplase represents a pre-formed streptokinase-plasminogen complex with plasminogen’s active site inactivated by anisoylation [245]. In vivo, p-anisic acid is cleaved and removed, resulting in an enzymatically active plasminogen activator. While keeping the original activity and antigenicity of streptokinase, the half-life of the variant was prolonged approximately 10-fold [246,247].
5. Staphylokinase
5.1. Biological Function
Staphylokinase is a small prokaryotic plasminogen activator secreted by lysogenic Staphylococcus aureus, enabling invasion of the host’s tissue [248]. Originally mentioned already during the discovery of streptokinase in 1933 [182], the molecule of staphylokinase was firstly described in 1948 [248] and produced recombinantly in 1983 [249]. Although being immunogenic and having the half-life of only 6 min, its high fibrin specificity, low production cost, and promising clot penetrability make staphylokinase a potential thrombolytic for the clinical practice [47,[250], [251], [252], [253]].
5.2. Molecular Structure
The protein has the smallest structure of all the biochemically characterized biological thrombolytics. It is composed of 136 amino acids, of which 30 % are charged [249,254,255]. The protein is folded into a single domain with the molecular weight of 15.5 kDa [256,257]. Structurally, but not sequentially, staphylokinase is homologous with the α domain of streptokinase with the β-grasp fold (Fig. 14) [256,258].
Fig. 14.
The complex of staphylokinase with bound plasmin. The complex of staphylokinase (gray) and the catalytic domain of plasmin (blue) (PDB ID 1BUI). The catalytic triad of plasmin His603, Asp646, and Ser741 are shown as red beads.
5.3. Mechanism of Activation and Specificity
Similarly to streptokinase, staphylokinase is an activator with an indirect mechanism of plasminogen activation, showing no enzymatic activity [248,259]. Compared to streptokinase, the complex of plasminogen bound by staphylokinase is inactive and is not able to perform the non-proteolytic activation of the plasminogen zymogen [260,261]. Instead, staphylokinase forms a productive equimolar (1:1) complex with plasmin only (Fig. 13 – Pathway II) [259,261,262]. Within the complex, staphylokinase modifies a broad substrate specificity of plasmin from a general protease digesting fibrin to an enzyme selectively cleaving the Arg561-Val562 bond in another molecule of plasminogen, yielding active plasmin [259]. Only a few studies have been conducted to understand staphylokinase at the molecular level. The results identified that the residues 26, 42–50, 65–69, and 75 are important for the plasmin “partner” binding [[263], [264], [265], [266], [267]], while the residues 11–16, 46–50, 65–69, and 97–98 are involved in the binding and the processing of the plasminogen “substrate” [256,266,[268], [269], [270]]. In contrast, the first ten N-terminal residues are not required for the activity and are cleaved by active plasmin [268,[271], [272], [273], [274], [275]].
5.4. Binding and Stimulation by Fibrin
An unusually high fibrin specificity of staphylokinase is given by mutual action of fibrin and α2-antiplasmin. In the absence of fibrin, the staphylokinase-plasmin complex is rapidly inhibited by α2-antiplasmin. In clotted plasma, fibrin competitively interacts with the complex via the same lysine binding sites as the inhibitor so the inhibition is prevented [[226], [227], [228],[276], [277], [278]]. Such a mechanism allows staphylokinase to activate plasminogen preferentially only on the fibrin’s surface. This avoids systemic plasminogen activation causing fibrinogenolysis and bleeding complications [226,228,253,276,277,279]. Moreover, the activity of staphylokinase is further enhanced by the fact that plasminogen/plasmin molecules bound to fibrin exhibit more extended conformation preferred by staphylokinase [278,280].
5.5. Inhibition
Compared to streptokinase, which is resistant to α2-antiplasmin in any form, the staphylokinase-plasmin complex is rapidly inhibited in human plasma as described above [227,228,276,277,279]. Conveniently, staphylokinase can reversibly dissociate from the inhibited complex so its effective concentration in plasma is not decreased by the inhibition [281]. The main factor decreasing the efficiency of staphylokinase is, therefore, its immunogenicity causing the production of neutralizing antibodies [250].
5.6. Protein Engineering
The immunogenic property of staphylokinase prevents its repeated administration and represents its biggest disadvantage. Three non-overlapping immunodominant regions located on the protein surface represent the main epitopes recognized by the antibodies [[282], [283], [284], [285]]. After disruption of the identified contacts by combinatorial mutagenesis, the immunogenicity of the best variant decreased to less than 30 % [283,[286], [287], [288], [289]]. Other engineering strategies using PEGylation [[290], [291], [292], [293], [294], [295]], glycosylation [296], protein fusion [288,[297], [298], [299], [300], [301]], and lipidification [302] provided variants of staphylokinase exhibiting higher efficiency, improved half-life, decreased immunogenicity, and a lower risk of reocclusion.
6. Desmoteplase
6.1. Biological Function
Desmoteplase is a plasminogen activator that was isolated from the saliva of the vampire bat Desmodus rotundus in 1974 [303]. It is employed by these hematophagous animals during feeding on the blood of livestock, maintaining blood fluidity and attenuating formation of clots. Thrombolytic properties of desmoteplase were observed already in 1932, but it took another 34 years to identify that this factor is an activator of plasminogen [304]. Thanks to its enormous fibrin specificity, an absence of neurotoxicity, and a highly prolonged half-life of more than 2 h, desmoteplase represents a promising activator with the potential for the treatment of cardiovascular diseases [116,[305], [306], [307], [308], [309]]. Although these beneficial properties were not observed during randomized clinical trials, further clinical tests are in progress [310,311].
6.2. Molecular Structure
The protein exhibits more than 72 % sequence homology with the human t-PA and analogically contains a finger F, an epidermal growth factor EGF, and a serine protease P domain (Fig. 15) but only one kringle K domain which is equivalent to kringle K1 in t-PA and lacks a lysine-binding site [[312], [313], [314]]. The structure is composed of 441 amino acids with the molecular weight of 52 kDa, contains three glycosylation sites Thr68, Asn117 and Asn362, and 14 disulfide bonds [312,313,[315], [316], [317]]. Desmoteplase does not possess any plasmin-sensitive cleavage site so it exists solely as a single chain form [314].
Fig. 15.
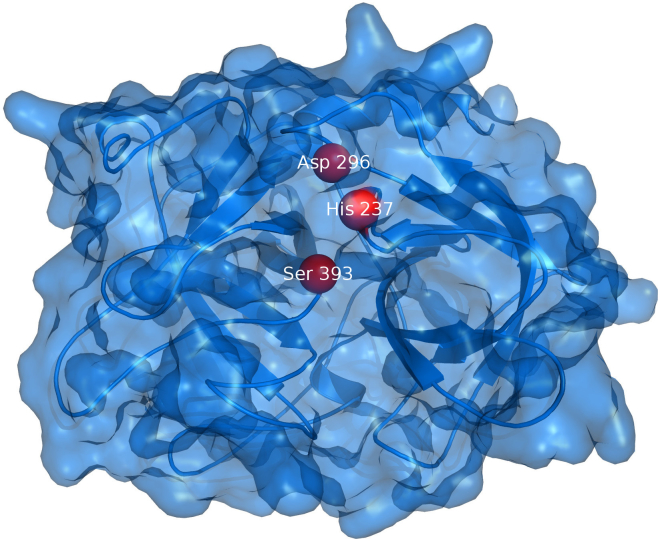
The catalytic domain of desmoteplase. The catalytic domain of desmoteplase (PDB ID 1A5I) from the vampire bat Desmodus rotundus is shown as blue surface. The catalytic triad of His237, Asp296, and Ser393 is shown as red beads.
6.3. Mechanism of Activation and Specificity
Desmoteplase is a serine protease with the typical catalytic triad (His238-Asp297-Ser394) that cleaves the Arg561-Val562 peptide bond in plasminogen, yielding active plasmin [[312], [313], [314]]. However, the mechanism of the enormous desmoteplase specificity has not been explained yet and all the experimental data provide only indirect assumptions [312,314,[318], [319], [320]]. It is hypothesized that the activity is connected with the intramolecular interaction between Lys259 and Asp297. This interaction opens the active site cavity and is formed only when desmoteplase is bound to fibrin by the finger F domain. Dissociation from fibrin disrupts this interaction and leads to a reversible loss of activity [314,318,320]. Such a mechanism also corresponds to the exclusive single-chain form of desmoteplase because the side chain of its Lys259 substitutes the function of the new N-terminus important in two-chain forms of other eukaryotic activators [314].
6.4. Binding and Stimulation by Fibrin
Desmoteplase is highly specific towards fibrin and exhibits the highest stimulation of all known thrombolytics [312,315,318,321]. Desmoteplase is almost inactive towards plasminogen in the absence of fibrin but once bound, its activity reaches the level of stimulated t-PA. As a consequence, desmoteplase is stimulated by fibrin approximately 100,000-fold, which is 200-fold higher than described for t-PA [312,318,321]. Moreover, the presence of fibrinogen and other plasma factors induce desmoteplase’s activity insignificantly so this activator is selective exclusively towards fibrin [318,319,321]. Such property prevents fibrinogen depletion and plasminemia which often cause side effects such as hemorrhage [305,306,308].
6.5. Inhibition
Despite desmoteplase’s eukaryotic origin, its repeated administration caused a slight raise of neutralizing antibodies in animal models, potentially decreasing the thrombolytic’s efficiency [322]. Furthermore, the structure is recognized by the inhibitors PAI-1 and PAI-2 due to its similarity to t-PA and urokinase, yet the resistance to inhibition is relatively high [306].
6.6. Protein Engineering
Since desmoteplase exhibits plenty of positive properties and not many drawbacks, a little effort was put into its engineering. A chimeric protein combining the structures of desmoteplase and tenecteplase has been recently reported [132,323]. In this variant, the kringle 2 domain of tenecteplase was deleted and the finger domain was exchanged for the domain of desmoteplase with the aim to improve the protein’s fibrin specificity. However, the constructed variant was only 8-fold more specific than t-PA but 25-fold less specific compared to desmoteplase [132]. Another approach was focused on preventing the creation of the two-chain form of t-PA. Substitution of the plasmin sensitive cleavage site with the corresponding sequence of desmoteplase (Arg275His, Ile276Ser, Lys277Thr) provided 28-fold higher fibrin specificity [318]. On the other hand, the reversed approach introducing the plasmin sensitive site (His191Arg, Ser192Ile, Thr193Lys) into desmoteplase led to the loss of specific activation which confirmed the importance of this region for the fibrin selectivity [318].
7. Perspectives
7.1. Novel Enzymes
Discovery of enzymes potentially serving as novel thrombolytics with interesting biological properties is a very attractive avenue for future research. Continuous advances in the next generation sequencing technologies provide a wealth of valuable information, which are stored in public genomic databases. These sequences can be systematically searched for novel biocatalysts by using bioinformatics tools and analyzed by computer modeling [324], cloned and experimentally characterized. Several enzymes directly cleaving fibrin, without plasminogen activation, have been recently described: (i) leech-derived Harobin, (ii) fibrinolytic enzyme from Chlorella algae and (iii) Nattokinase from soybeans [[325], [326], [327]]. The hunt for novel thrombolytics will result in the discovery of more potent enzymes with different mechanisms than the ones known to date.
7.2. Small Molecules
Small molecules from a marine fungus Stachybotrys such as staplabin bind and induce a conformational change in plasminogen, increasing its fibrin binding and susceptibility to activation by plasminogen activators [328]. Additional molecules are being isolated from different organisms or developed by chemical modification since the discovery of staplabin [329,330]. Their small size confers several advantages as easier production, lack of immunogenicity, anti-inflammatory effects and no cross-reactivity with other pathways [327]. Despite their small size, these molecules exhibit clot-targeted plasminogen activation [331] and current animal studies have shown promising results for example with the molecule SMTP-7 [332,333]. Isolation and chemical modification of novel biologically active small compounds is another promising trend in thrombolytics research.
7.3. Encapsulation & Targeted Release
Plasminogen activators’ half-lives can be significantly enhanced by trapping them in liposomes and nanomaterials. Effectiveness of plasminogen activators can be improved by targeting their release using ultrasound, the presence of activated platelets, or shear-stress [[334], [335], [336], [337], [338]]. Encapsulation allows for the targeted release of both a neuroprotective and a thrombolytic agent [339]. Modification with covalently linked DNA or RNA allows disruption of interactions between t-PA and LRP1 receptor with concurrent regulation of enzymatic activity [340,341]. PEGylation can reduce immunogenicity and increase the half-life of prokaryotic plasminogen activators [239,292]. Newly established methods for targeted delivery will be used to target the action of plasminogen activators into the thrombus with minimized side-effects.
7.4. Adjuvants
Neurological side-effects such as neuroinflammation and disruption of the blood-brain barrier by the interaction of various molecules with t-PA and plasmin negatively influence the outcome of the therapy. Unfavorable interactions and oxidative stress on thrombolytics in ischemic conditions are being tackled by using antibodies, protein antagonists of receptor binding, or small molecules [[342], [343], [344], [345], [346], [347], [348]]. Anti-inflammatory cytokines were recently tested as adjuvant therapy to alleviate inflammatory side-effects [348,349]. Plasmin and plasminogen activators are inhibited quickly in the blood clot. Moreover, binding sites of plasminogen activators on fibrin are removed by tissue activable fibrinolysis inhibitor which also slows down fibrinolysis. These effects can be blocked via antibodies against PAI-1, α2-antiplasmin, and tissue activatable fibrinolysis inhibitor, or by blocking two fibrinolytic inhibitors simultaneously by diabodies. This has proven to accelerate fibrinolysis in vivo [[350], [351], [352], [353], [354], [355], [356]]. PAI-1 can be also inhibited by small molecules such as tiplaxtinin or downregulated by statins and renin-angiotensin-aldosterone inhibitors [357]. The fibrinolytic system has complex connections to immune pathways, cerebrovascular coupling, nervous system, cell migration, metastasis, and cognition. These connections offer many targets for enhancing the efficacy of thrombolytics as well as their use for the treatment of non-thrombotic conditions.
7.5. Protein Engineering
Protein engineering studies of eukaryotic plasminogen activators are no longer aimed at increasing half-life since longer-circulating and inhibitor-resistant thrombolytics are available. One of the aims is reducing the side-effects via increasing fibrin specificity [358,359] or preventing interactions with the receptors [114,115,360]. Reducing side-effects also provides novel indications for thrombolytic therapy, such as drainage of intracerebral hemmorhages or extension of the time window in which thrombolytics can be administered [114,333]. Despite improvements in many key properties, e.g., fibrin selectivity, half-life, and inhibitor resistance; faster lysis of a thrombus is still of major concern. The structure-function relationships of fibrin-specific drugs need to be studied in a complex with fibrin, a large molecule of its own. The improvement in the structural techniques, such as small-angle X-ray scattering and cryo-EM, will allow solving even the large multi-protein complexes. Acquired structural data can be analyzed by various structural bioinformatics and computer modeling techniques that will bring insights for engineering activity, affinity, specificity and selectivity of thrombolytics.
7.6. Production Systems
Seventeen disulfide bridges of t-PA make this protein hard to express in prokaryotic systems. Even reteplase containing nine disulfide bridges have to be refolded after the purification in order to adopt an active conformation. Higher yields of properly folded t-PA were achieved via expression into the periplasm or co-expression with disulfide oxidoreductase [361,362]. Production in yeasts generally leads to higher yields and absence of endotoxin when compared to bacteria [[363], [364], [365]]. Other avenues are auto-induction, growth medium or cultivation protocol optimization [366,367], development of new refolding protocols [[368], [369], [370]] or new mammalian cell lines [371]. To avoid the demanding work with mammalian cell cultures, eukaryotic plasminogen activators can be produced in plants or extracted from the milk of transgenic animals [[372], [373], [374], [375], [376]]. We envisage fine-tuning of available production systems and continuous search for the new expression and purification systems.
Acknowledgements
The authors would like to express their thanks to the Czech Ministry of Education (CZ.02.1.01/0.0/0.0/16_026/0008451, CZ.02.1.01/0.0/0.0/16_013/0001761, LM2015047) and Technology Agency of the Czech Republic (TN01000013) for financial support. CERIT-SC and META are acknowledged for providing the computational resources for our modeling work (LM2015042, LM2015085). Martin Toul greatly acknowledges the financial support of his doctoral study by the scholarship Brno Ph.D. Talent.
Declarations of Competing Interests
The authors declare that they have no conflict of interest.
Contributor Information
David Bednar, Email: 222755@mail.muni.cz.
Jiri Damborsky, Email: jiri@chemi.muni.cz.
References
- 1.TPJ Bryan, 18 J The rise and fall of the clot buster: a review on the history of streptokinase. 2014. http://www.pharmaceutical-journal.com/news-and-analysis/features/the-rise-and-fall-of-the-clot-buster-a-review-on-the-history-of-streptokinase/20065679.article Pharm J. (accessed September 22, 2017)
- 2.Proctor P., Leesar M.A., Chatterjee A. Thrombolytic therapy in the current ERA: myocardial infarction and beyond. Curr Pharm Des. 2018;24:414–426. doi: 10.2174/1381612824666171227211623. [DOI] [PubMed] [Google Scholar]
- 3.Niego B., Freeman R., Puschmann T.B., Turnley A.M. Medcalf RL. t-PA–specific modulation of a human blood-brain barrier model involves plasmin-mediated activation of the Rho kinase pathway in astrocytes. Blood. 2012;119:4752–4761. doi: 10.1182/blood-2011-07-369512. [DOI] [PubMed] [Google Scholar]
- 4.Lesept F., Chevilley A., Jezequel J., Ladepeche L., Macrez R., Aimable M. Tissue-type plasminogen activator controls neuronal death by raising surface dynamics of extrasynaptic NMDA receptors. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehra A., Ali C., Parcq J., Vivien D., Docagne F. The plasminogen activation system in neuroinflammation. Biochim Biophys Acta BBA Mol Basis Dis. 1862;2016:395–402. doi: 10.1016/j.bbadis.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Oh S.B., Byun C.J., Yun J.-H., Jo D.-G., Carmeliet P., Koh J.-Y. Tissue plasminogen activator arrests Alzheimer’s disease pathogenesis. Neurobiol Aging. 2014;35:511–519. doi: 10.1016/j.neurobiolaging.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Akhter H., Huang W.-T., van Groen T., Kuo H.-C., Miyata T., Liu R.-M. A small molecule inhibitor of plasminogen activator inhibitor-1 Reduces brain amyloid-β load and improves memory in an animal model of alzheimer’s disease. J Alzheimers Dis. 2018;64:447–457. doi: 10.3233/JAD-180241. [DOI] [PubMed] [Google Scholar]
- 8.Tsai S.-J. Role of tissue-type plasminogen activator and plasminogen activator inhibitor-1 in psychological stress and depression. Oncotarget. 2017;8:113258–113268. doi: 10.18632/oncotarget.19935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyganowska-Świątkowska M., Wyganowska-Świątkowska M., Tarnowski M., Tarnowski M., Murtagh D., Murtagh D. Proteolysis is the most fundamental property of malignancy and its inhibition may be used therapeutically (Review) Int J Mol Med. 2019;43:15–25. doi: 10.3892/ijmm.2018.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoylaerts M., Rijken D.C., Lijnen H.R., Collen D. Kinetics of the activation of plasminogen by human tissue plasminogen activator. Role of fibrin J Biol Chem. 1982;257:2912–2919. [PubMed] [Google Scholar]
- 11.Wun T.C., Schleuning W.D., Reich E. Isolation and characterization of urokinase from human plasma. J Biol Chem. 1982;257:3276–3283. [PubMed] [Google Scholar]
- 12.Khasa Y.P. The evolution of recombinant thrombolytics: current status and future directions. Bioengineered. 2016;8:331–358. doi: 10.1080/21655979.2016.1229718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsgren M., Råden B., Israelsson M., Larsson K., Hedén L.-O. Molecular cloning and characterization of a full-length cDNA clone for human plasminogen. FEBS Lett. 1987;213:254–260. doi: 10.1016/0014-5793(87)81501-6. [DOI] [PubMed] [Google Scholar]
- 14.Torrèns I., Ojalvo A.G., Seralena A., Hayes O., de la Fuente J. A mutant streptokinase lacking the C-terminal 42 amino acids is less immunogenic. Immunol Lett. 2000;70:213–218. doi: 10.1016/s0165-2478(99)00151-0. [DOI] [PubMed] [Google Scholar]
- 15.Castellino F., Ploplis V. Structure and function of the plasminogen/plasmin system. Thromb Haemost. 2005;93:647–654. doi: 10.1160/TH04-12-0842. [DOI] [PubMed] [Google Scholar]
- 16.Wang H., Prorok M., Bretthauer R.K., Castellino F.J. Serine-578 is a major phosphorylation locus in human plasma plasminogen †. Biochemistry. 1997;36:8100–8106. doi: 10.1021/bi970328d. [DOI] [PubMed] [Google Scholar]
- 17.Wu G., Quek A.J., Caradoc-Davies T.T., Ekkel S.M., Mazzitelli B., Whisstock J.C. Structural studies of plasmin inhibition. Biochem Soc Trans. 2019;47:541–557. doi: 10.1042/BST20180211. [DOI] [PubMed] [Google Scholar]
- 18.Robbins K.C., Summaria L., Hsieh B., Shah R.J. The peptide chains of human plasmin. mechanism of activation of human plasminogen to plasmin. J Biol Chem. 1967;242:2333–2342. [PubMed] [Google Scholar]
- 19.Suenson E., Thorsen S. Secondary-site binding of Glu-plasmin, Lys-plasmin and miniplasmin to fibrin. Biochem J. 1981;197:619–628. doi: 10.1042/bj1970619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miles L.A., Dahlberg C.M., Plow E.F. The cell-binding domains of plasminogen and their function in plasma. J Biol Chem. 1988;263:11928–11934. [PubMed] [Google Scholar]
- 21.Xue Y., Bodin C., Olsson K. Crystal structure of the native plasminogen reveals an activation-resistant compact conformation. J Thromb Haemost. 2012;10:1385–1396. doi: 10.1111/j.1538-7836.2012.04765.x. [DOI] [PubMed] [Google Scholar]
- 22.Fredenburgh J.C., Nesheim M.E. Lys-plasminogen is a significant intermediate in the activation of Glu-plasminogen during fibrinolysis in vitro. J Biol Chem. 1992;267:26150–26156. [PubMed] [Google Scholar]
- 23.Law R.H.P., Caradoc-Davies T., Cowieson N., Horvath A.J., Quek A.J., Encarnacao J.A. The X-ray crystal structure of full-length human plasminogen. Cell Rep. 2012;1:185–190. doi: 10.1016/j.celrep.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Oh C.-W., Hoover-Plow J., Plow E.F. The role of plasminogen in angiogenesis in vivo. J Thromb Haemost. 2003;1:1683–1687. doi: 10.1046/j.1538-7836.2003.00182.x. [DOI] [PubMed] [Google Scholar]
- 25.Moser T., Stack M., Wahl M., Pizzo S. The mechanism of action of angiostatin: can you teach an old dog new tricks? Thromb Haemost. 2002;87:394–401. [PubMed] [Google Scholar]
- 26.Geiger J.H., Cnudde S.E. What the structure of angiostatin may tell us about its mechanism of action. J Thromb Haemost. 2004;2:23–34. doi: 10.1111/j.1538-7836.2004.00544.x. [DOI] [PubMed] [Google Scholar]
- 27.Wahl M.L., Kenan D.J., Gonzalez-Gronow M., Pizzo S.V. Angiostatin’s molecular mechanism: aspects of specificity and regulation elucidated. J Cell Biochem. 2005;96:242–261. doi: 10.1002/jcb.20480. [DOI] [PubMed] [Google Scholar]
- 28.Abad M.C., Arni R.K., Grella D.K., Castellino F.J., Tulinsky A., Geiger J.H. The X-ray crystallographic structure of the angiogenesis inhibitor angiostatin. J Mol Biol. 2002;318:1009–1017. doi: 10.1016/S0022-2836(02)00211-5. [DOI] [PubMed] [Google Scholar]
- 29.Thiebaut A.M., Gauberti M., Ali C., Martinez De Lizarrondo S., Vivien D., Yepes M. The role of plasminogen activators in stroke treatment: fibrinolysis and beyond. Lancet Neurol. 2018;17:1121–1132. doi: 10.1016/S1474-4422(18)30323-5. [DOI] [PubMed] [Google Scholar]
- 30.Diamond S.L. Engineering design of optimal strategies for blood clot dissolution. Annu Rev Biomed Eng. 1999;1:427. doi: 10.1146/annurev.bioeng.1.1.427. [DOI] [PubMed] [Google Scholar]
- 31.Marcos-Contreras O.A., Ganguly K., Yamamoto A., Shlansky-Goldberg R., Cines D.B., Muzykantov V.R. Clot penetration and retention by plasminogen activators promote fibrinolysis. Biochem Pharmacol. 2013;85:216–222. doi: 10.1016/j.bcp.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 32.Bannish B.E., Chernysh I.N., Keener J.P., Fogelson A.L., Weisel J.W. Molecular and physical mechanisms of fibrinolysis and thrombolysis from mathematical modeling and experiments. Sci Rep. 2017;7 doi: 10.1038/s41598-017-06383-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mutch N.J., Thomas L., Moore N.R., Lisiak K.M., Booth N.A. TAFIa, PAI-1 and α2-antiplasmin: complementary roles in regulating lysis of thrombi and plasma clots. J Thromb Haemost. 2007;5:812–817. doi: 10.1111/j.1538-7836.2007.02430.x. [DOI] [PubMed] [Google Scholar]
- 34.Krishnamurti C., Keyt B., Maglasang P., Alving B.M. PAI-1-resistant t-PA: low doses prevent fibrin deposition in rabbits with increased PAI-1 activity. Blood. 1996;87:14–19. [PubMed] [Google Scholar]
- 35.Kheiri B., Osman M., Abdalla A., Haykal T., Ahmed S., Hassan M. Tenecteplase versus alteplase for management of acute ischemic stroke: a pairwise and network meta-analysis of randomized clinical trials. J Thromb Thrombolysis. 2018;46:440–450. doi: 10.1007/s11239-018-1721-3. [DOI] [PubMed] [Google Scholar]
- 36.Bode C., Smalling Richard W., Berg G., Burnett C., Lorch G., Kalbfleisch John M. Randomized comparison of coronary thrombolysis achieved with double-bolus reteplase (recombinant plasminogen activator) and front-loaded, accelerated alteplase (recombinant tissue plasminogen activator) in patients with acute myocardial infarction. Circulation. 1996;94:891–898. doi: 10.1161/01.cir.94.5.891. [DOI] [PubMed] [Google Scholar]
- 37.Smalling Richard W., Bode C., Kalbfleisch J., Sen S., Limbourg P., Forycki F. More rapid, Complete, and stable coronary thrombolysis with bolus administration of reteplase compared with alteplase infusion in acute myocardial infarction. Circulation. 1995;91:2725–2732. doi: 10.1161/01.cir.91.11.2725. [DOI] [PubMed] [Google Scholar]
- 38.Campbell B.C.V., Mitchell P.J., Churilov L., Yassi N., Kleinig T.J., Dowling R.J. Tenecteplase versus alteplase before thrombectomy for ischemic stroke. N Engl J Med. 2018;10 doi: 10.1056/NEJMoa1716405. [DOI] [PubMed] [Google Scholar]
- 39.Xu N., Chen Z., Zhao C., Xue T., Wu X., Sun X. Different doses of tenecteplase vs alteplase in thrombolysis therapy of acute ischemic stroke: evidence from randomized controlled trials. Drug Des Devel Ther. 2018;12:2071–2084. doi: 10.2147/DDDT.S170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X., Ling L., Li C., Ma Q. Efficacy and safety of desmoteplase in acute ischemic stroke patients a systematic review and meta-analysis. Medicine (Baltimore) 2017;96 doi: 10.1097/MD.0000000000006667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verheugt F.W.A., Meijer A., Lagrand W.K., van Eenige M.J. Reocclusion: the flip side of coronary thrombolysis. J Am Coll Cardiol. 1996;27:766–773. doi: 10.1016/0735-1097(95)00492-0. [DOI] [PubMed] [Google Scholar]
- 42.Arterial reocclusion in stroke patients treated with intravenous tissue plasminogen activatorCurr Med Lit Stroke Rev. 2003;7:56. doi: 10.1212/wnl.59.6.862. [DOI] [PubMed] [Google Scholar]
- 43.Parcq J., Bertrand T., Montagne A., Baron A.F., Macrez R., Billard J.M. Unveiling an exceptional zymogen: the single-chain form of tPA is a selective activator of NMDA receptor-dependent signaling and neurotoxicity. Cell Death Differ. 2012;19:1983–1991. doi: 10.1038/cdd.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonias S.L., Banki M.A., Gilder A.S., Campana W.M., Mantuano E. PAI1 blocks effects of tissue-type plasminogen activator on cell-signaling and physiology mediated by the NMDA receptor. J Cell Sci. 2018;2018:37. doi: 10.1242/jcs.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fredriksson L., Lawrence D.A., Medcalf R.L. tPA modulation of the blood–brain barrier: a unifying explanation for the pleiotropic effects of tPA in the CNS. Semin Thromb Hemost. 2017;43:154–168. doi: 10.1055/s-0036-1586229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.del Zoppo G.J., Saver J.L., Jauch E.C., Adams H.P. Expansion of the time window for treatment of acute ischemic stroke with intravenous tissue plasminogen activator a science advisory from the american heart association/american stroke association. Stroke. 2009;40:2945–2948. doi: 10.1161/STROKEAHA.109.192535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baruah D.B., Dash R.N., Chaudhari M.R., Kadam S.S. Plasminogen activators: a comparison. Vascul Pharmacol. 2006;44:1–9. doi: 10.1016/j.vph.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Gurewich V. Therapeutic fibrinolysis: how efficacy and safety can be improved. J Am Coll Cardiol. 2016;68:2099–2106. doi: 10.1016/j.jacc.2016.07.780. [DOI] [PubMed] [Google Scholar]
- 49.Medcalf R.L. Fibrinolysis: from blood to the brain. J Thromb Haemost. 2017;15:2089–2098. doi: 10.1111/jth.13849. [DOI] [PubMed] [Google Scholar]
- 50.Benchenane K., Berezowski V., Fernández-Monreal M., Brillault J., Valable S., Dehouck M.-P. Oxygen glucose deprivation switches the transport of tPA across the blood–brain barrier from an LRP-dependent to an increased LRP-independent process. Stroke. 2005;36:1059–1064. doi: 10.1161/01.STR.0000163050.39122.4f. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki Y., Nagai N., Umemura K. A review of the mechanisms of blood-brain barrier permeability by tissue-type plasminogen activator treatment for cerebral ischemia. Front Cell Neurosci. 2016;10:2. doi: 10.3389/fncel.2016.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y., Li D., Zhao J., Song J., Zhao Y. The role of the low-density lipoprotein receptor-related protein 1 (LRP-1) in regulating blood-brain barrier integrity. Rev Neurosci Berl. 2016;27:623–634. doi: 10.1515/revneuro-2015-0069. [DOI] [PubMed] [Google Scholar]
- 53.Ishiguro M., Kawasaki K., Suzuki Y., Ishizuka F., Mishiro K., Egashira Y. A Rho kinase (ROCK) inhibitor, fasudil, prevents matrix metalloproteinase-9-related hemorrhagic transformation in mice treated with tissue plasminogen activator. Neuroscience. 2012;220:302–312. doi: 10.1016/j.neuroscience.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 54.Nassar T. In vitro and in vivo effects of tPA and PAI-1 on blood vessel tone. Blood. 2003;103:897–902. doi: 10.1182/blood-2003-05-1685. [DOI] [PubMed] [Google Scholar]
- 55.Cole E.S., Nichols E.H., Poisson L., Harnois M.L., Livingston D.J. In vivo clearance of tissue plasminogen activator: the complex role of sites of glycosylation and level of sialylation. Fibrinolysis. 1993;7:15–22. [Google Scholar]
- 56.Ny T., Elgh F., Lund B. The structure of the human tissue-type plasminogen activator gene: correlation of intron and exon structures to functional and structural domains. Proc Natl Acad Sci U S A. 1984;81:5355–5359. doi: 10.1073/pnas.81.17.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verheijen J.H., Caspers M.P., Chang G.T., de Munk G.A., Pouwels P.H., Enger-Valk B.E. Involvement of finger domain and kringle 2 domain of tissue-type plasminogen activator in fibrin binding and stimulation of activity by fibrin. EMBO J. 1986;5:3525–3530. doi: 10.1002/j.1460-2075.1986.tb04678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beebe D.P., Miles L.A., Plow E.F. A linear amino acid sequence involved in the interaction of t-PA with its endothelial cell receptor. Blood. 1989;74:2034–2037. [PubMed] [Google Scholar]
- 59.Camani C., Kruithof E.K. The role of the finger and growth factor domains in the clearance of tissue-type plasminogen activator by hepatocytes. J Biol Chem. 1995;270:26053–26056. doi: 10.1074/jbc.270.44.26053. [DOI] [PubMed] [Google Scholar]
- 60.Correa F., Gauberti M., Parcq J., Macrez R., Hommet Y., Obiang P. Tissue plasminogen activator prevents white matter damage following stroke. J Exp Med. 2011;208:1229–1242. doi: 10.1084/jem.20101880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harris R.J., Leonard C.K., Guzzetta A.W., Spellman M.W. Tissue plasminogen activator has an O-linked fucose attached to threonine-61 in the epidermal growth factor domain. Biochemistry. 1991;30:2311–2314. doi: 10.1021/bi00223a004. [DOI] [PubMed] [Google Scholar]
- 62.Narita M., Bu G., Herz J., Schwartz A.L. Two receptor systems are involved in the plasma clearance of tissue-type plasminogen activator (t-PA) in vivo. J Clin Invest. 1995;96:1164–1168. doi: 10.1172/JCI118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gething M.J., Adler B., Boose J.A., Gerard R.D., Madison E.L., McGookey D. Variants of human tissue-type plasminogen activator that lack specific structural domains of the heavy chain. EMBO J. 1988;7:2731–2740. doi: 10.1002/j.1460-2075.1988.tb03127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rehan M., Sagar A., Sharma V., Mishra S., Ashish Sahni G. Penta-l-lysine potentiates fibrin-independent activity of human tissue plasminogen activator. J Phys Chem B. 2015;119:13271–13277. doi: 10.1021/acs.jpcb.5b07735. [DOI] [PubMed] [Google Scholar]
- 65.De Munk G.A.W., Caspers M.P.M., Chang G.T.G., Pouwels P.H., Enger-Valk B.E., Verheijen J.H. Binding of tissue-type plasminogen activator to lysine, lysine analogs, and fibrin fragments. Biochemistry. 1989;28:7318–7325. doi: 10.1021/bi00444a026. [DOI] [PubMed] [Google Scholar]
- 66.Kranenburg O., Bouma B., Kroon-Batenburg L.M.J., Reijerkerk A., Wu Y.-P., Voest E.E. Tissue-type plasminogen activator is a multiligand cross-β structure receptor. Curr Biol. 2002;12:1833–1839. doi: 10.1016/s0960-9822(02)01224-1. [DOI] [PubMed] [Google Scholar]
- 67.Parcq J., Bertrand T., Baron A.F., Hommet Y., Angles-Cano E., Vivien D. Molecular requirements for safer generation of thrombolytics by bioengineering the tissue-type plasminogen activator A chain. J Thromb Haemost. 2013;11:539–546. doi: 10.1111/jth.12128. [DOI] [PubMed] [Google Scholar]
- 68.Aisina R.B., Mukhametova L.I. Structure and function of plasminogen/plasmin system. Russ J Bioorganic Chem. 2014;40:590–605. [Google Scholar]
- 69.Lamba D., Bauer M., Huber R., Fischer S., Rudolph R., Kohnert U. The 2.3 Å crystal structure of the catalytic domain of recombinant two-chain human tissue-type plasminogen activator. J Mol Biol. 1996;258:117–135. doi: 10.1006/jmbi.1996.0238. [DOI] [PubMed] [Google Scholar]
- 70.Tachias K., Madison E.L. Converting tissue type plasminogen activator into a zymogen. important role of Lys156. J Biol Chem. 1997;272:28–31. doi: 10.1074/jbc.272.1.28. [DOI] [PubMed] [Google Scholar]
- 71.Holmes W.E., Pennica D., Blaber M., Rey M.W., Guenzler W.A., Steffens G.J. Cloning and expression of the gene for pro-urokinase in Escherichia coli. Bio/Technology. 1985;3:923. [Google Scholar]
- 72.Bode W., Renatus M. Tissue-type plasminogen activator: variants and crystal/solution structures demarcate structural determinants of function. Curr Opin Struct Biol. 1997;7:865–872. doi: 10.1016/s0959-440x(97)80159-5. [DOI] [PubMed] [Google Scholar]
- 73.Vogt A.D., Chakraborty P., Di Cera E. Kinetic dissection of the pre-existing conformational equilibrium in the trypsin fold. J Biol Chem. 2015;290:22435–22445. doi: 10.1074/jbc.M115.675538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chakraborty P., Acquasaliente L., Pelc L.A., Di Cera E. Interplay between conformational selection and zymogen activation. Sci Rep. 2018;8:4080. doi: 10.1038/s41598-018-21728-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coombs G.S., Dang A.T., Madison E.L., Corey D.R. Distinct mechanisms contribute to stringent substrate specificity of tissue-type plasminogen activator. J Biol Chem. 1996;271:4461–4467. doi: 10.1074/jbc.271.8.4461. [DOI] [PubMed] [Google Scholar]
- 76.Ke S.H., Tachias K., Lamba D., Bode W., Madison E.L. Identification of a hydrophobic exosite on tissue type plasminogen activator that modulates specificity for plasminogen. J Biol Chem. 1997;272:1811–1816. doi: 10.1074/jbc.272.3.1811. [DOI] [PubMed] [Google Scholar]
- 77.Tachias K., Madison E. Variants of tissue-type plasminogen-activator which display substantially enhanced stimulation by fibrin. J Biol Chem. 1995;270:18319–18322. doi: 10.1074/jbc.270.31.18319. [DOI] [PubMed] [Google Scholar]
- 78.Rathore Y.S., Rehan M., Pandey K., Sahni G., Ashish First structural model of full-length human tissue-plasminogen activator: a SAXS data-based modeling study. J Phys Chem B. 2012;116:496–502. doi: 10.1021/jp207243n. [DOI] [PubMed] [Google Scholar]
- 79.Bakker A.H.F., Weening-Verhoeff E.J.D., Verheijen J.H. The role of the lysyl binding site of tissue-type plasminogen activator in the interaction with a forming fibrin clot. J Biol Chem. 1995;270:12355–12360. doi: 10.1074/jbc.270.21.12355. [DOI] [PubMed] [Google Scholar]
- 80.van Zonneveld A.J., Veerman H., Pannekoek H. On the interaction of the finger and the kringle-2 domain of tissue-type plasminogen activator with fibrin. inhibition of kringle-2 binding to fibrin by epsilon-amino caproic acid. J Biol Chem. 1986;261:14214–14218. [PubMed] [Google Scholar]
- 81.Paoni N.F., Chow A.M., Peña L.C., Keyt B.A., Zoller M.J., Bennett W.F. Making tissue-type plasminogen activator more fibrin specific. Protein Eng Des Sel. 1993;6:529–534. doi: 10.1093/protein/6.5.529. [DOI] [PubMed] [Google Scholar]
- 82.Lijnen H.R. Elements of the fibrinolytic system. In: Nieuwenhuizen W., Mosesson M.W., DeMaat M.P.M., editors. Fibrinogen. Vol. 936. New York Acad Sciences; New York: 2001. pp. 226–236. [DOI] [PubMed] [Google Scholar]
- 83.Medved L., Nieuwenhuizen W. Molecular mechanisms of initiation of fibrinolysis by fibrin. Thromb Haemost. 2003;89:409–419. [PubMed] [Google Scholar]
- 84.Tsurupa G., Medved L. Identification and characterization of novel tPA- and plasminogen-binding sites within fibrin(ogen) αC-domains †. Biochemistry. 2001;40:801–808. doi: 10.1021/bi001789t. [DOI] [PubMed] [Google Scholar]
- 85.Schielen W.J., Adams H.P., van Leuven K., Voskuilen M., Tesser G.I., Nieuwenhuizen W. The sequence gamma-(312-324) is a fibrin-specific epitope. Blood. 1991;77:2169–2173. [PubMed] [Google Scholar]
- 86.Voskuilen M., Vermond A., Veeneman G.H., van Boom J.H., Klasen E.A., Zegers N.D. Fibrinogen lysine residue A alpha 157 plays a crucial role in the fibrin-induced acceleration of plasminogen activation, catalyzed by tissue-type plasminogen activator. J Biol Chem. 1987;262:5944–5946. [PubMed] [Google Scholar]
- 87.Yakovlev S., Makogonenko E., Kurochkina N., Nieuwenhuizen W., Ingham K., Medved L. Conversion of fibrinogen to fibrin: mechanism of exposure of tPA- and plasminogen-binding sites. Biochemistry. 2000;39:15730–15741. doi: 10.1021/bi001847a. [DOI] [PubMed] [Google Scholar]
- 88.Doolittle R.F., Pandi L. Binding of synthetic B knobs to fibrinogen changes the character of fibrin and inhibits its ability to activate tissue plasminogen activator and its destruction by plasmin. Biochemistry. 2006;45:2657–2667. doi: 10.1021/bi0524767. [DOI] [PubMed] [Google Scholar]
- 89.Fleury V., Angles-Cano E. 2002. Characterization of the binding of plasminogen to fibrin surfaces: the role of carboxy-terminal lysines. [DOI] [PubMed] [Google Scholar]
- 90.Higgins D.L., Vehar G.A. Interaction of one-chain and two-chain tissue plasminogen activator with intact and plasmin-degraded fibrin. Biochemistry. 1987;26:7786–7791. doi: 10.1021/bi00398a038. [DOI] [PubMed] [Google Scholar]
- 91.Scott R.W., Bergman B.L., Bajpai A., Hersh R.T., Rodriguez H., Jones B.N. Protease nexin. Properties and a modified purification procedure. J Biol Chem. 1985;260:7029–7034. [PubMed] [Google Scholar]
- 92.Gardiner E.E., Medcalf R.L. Is plasminogen activator inhibitor type 2 really a plasminogen activator inhibitor after all? J Thromb Haemost. 2014;12:1703–1705. doi: 10.1111/jth.12704. [DOI] [PubMed] [Google Scholar]
- 93.Yepes M., Lawrence D. Neuroserpin: a selective inhibitor of tissue-type plasminogen activator in the central nervous system. Thromb Haemost. 2004;91:457–464. doi: 10.1160/TH03-12-0766. [DOI] [PubMed] [Google Scholar]
- 94.Rau J.C., Beaulieu L.M., Huntington J.A., Church F.C. Serpins in thrombosis, hemostasis and fibrinolysis. J Thromb Haemost. 2007;5:102–115. doi: 10.1111/j.1538-7836.2007.02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Madison E.L., Goldsmith E.J., Gerard R.D., Gething M.J., Sambrook J.F., Bassel-Duby R.S. Amino acid residues that affect interaction of tissue-type plasminogen activator with plasminogen activator inhibitor 1. Proc Natl Acad Sci U S A. 1990;87:3530–3533. doi: 10.1073/pnas.87.9.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gong L., Liu M., Zeng T., Shi X., Yuan C., Andreasen P.A. Crystal structure of the michaelis complex between tissue-type plasminogen activator and plasminogen activators inhibitor-1. J Biol Chem. 2015;290:25795–25804. doi: 10.1074/jbc.M115.677567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Keyt B.A., Paoni N.F., Refino C.J., Berleau L., Nguyen H., Chow A. A faster-acting and more potent form of tissue plasminogen activator. Proc Natl Acad Sci U S A. 1994;91:3670–3674. doi: 10.1073/pnas.91.9.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tachias K., Madison E.L. Variants of tissue-type plasminogen activator that display extraordinary resistance to inhibition by the serpin plasminogen activator inhibitor type 1. J Biol Chem. 1997;272:14580–14585. doi: 10.1074/jbc.272.23.14580. [DOI] [PubMed] [Google Scholar]
- 99.Vindigni A., Winfield M., Ayala Y.M., Di Cera E. Role of residue Y99 in tissue plasminogen activator. Protein Sci Publ Protein Soc. 2000;9:619–622. doi: 10.1110/ps.9.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Peng S., Xue G., Chen S., Chen Z., Yuan C., Li J. tPA point mutation at autolysis loop enhances resistance to PAI-1 inhibition and catalytic activity. Thromb Haemost. 2018;119:077–086. doi: 10.1055/s-0038-1676518. [DOI] [PubMed] [Google Scholar]
- 101.Horn I., van den Berg B., Moestrup S., Pannekoek H., van Zonneveld A.-J. Plasminogen activator inhibitor 1 contains a cryptic high affinity receptor binding site that is exposed upon complex formation with tissue-type plasminogen activator. Thromb Haemost. 1998;80:822–828. [PubMed] [Google Scholar]
- 102.Nagaoka M.R., Strital E., Kouyoumdjian M., Borges D.R. Participation of a galectin-dependent mechanism in the hepatic clearance of tissue-type plasminogen activator and plasma kallikrein. Thromb Res. 2002;108:257–262. doi: 10.1016/s0049-3848(02)00393-6. [DOI] [PubMed] [Google Scholar]
- 103.Willnow T.E., Goldstein J.L., Orth K., Brown M.S., Herz J. Low density lipoprotein receptor-related protein and gp330 bind similar ligands, including plasminogen activator-inhibitor complexes and lactoferrin, an inhibitor of chylomicron remnant clearance. J Biol Chem. 1992;267:26172–26180. [PubMed] [Google Scholar]
- 104.Horn I.R., van den Berg B.M., van der Meijden P.Z., Pannekoek H., van Zonneveld A.J. Molecular analysis of ligand binding to the second cluster of complement-type repeats of the low density lipoprotein receptor-related protein. evidence for an allosteric component in receptor-associated protein-mediated inhibition of ligand binding. J Biol Chem. 1997;272:13608–13613. doi: 10.1074/jbc.272.21.13608. [DOI] [PubMed] [Google Scholar]
- 105.Willnow T.E., Orth K., Herz J. Molecular dissection of ligand binding sites on the low density lipoprotein receptor-related protein. J Biol Chem. 1994;269:15827–15832. [PubMed] [Google Scholar]
- 106.Neels J.G., van den Berg B.M.M., Lookene A., Olivecrona G., Pannekoek H., van Zonneveld A.-J. The second and fourth cluster of class A cysteine-rich repeats of the low density lipoprotein receptor-related protein share ligand-binding properties. J Biol Chem. 1999;274:31305–31311. doi: 10.1074/jbc.274.44.31305. [DOI] [PubMed] [Google Scholar]
- 107.Fisher C., Beglova N., Blacklow S.C. Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol Cell. 2006;22:277–283. doi: 10.1016/j.molcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 108.Guttman M., Prieto J.H., Handel T.M., Domaille P.J., Komives E.A. Structure of the Minimal interface between ApoE and LRP. J Mol Biol. 2010;398:306–319. doi: 10.1016/j.jmb.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gettins P.G.W., Dolmer K. The high affinity binding site on plasminogen activator inhibitor-1 (PAI-1) for the low density lipoprotein receptor-related protein (LRP1) Is composed of four basic residues. J Biol Chem. 2016;291:800–812. doi: 10.1074/jbc.M115.688820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dolmer K., Campos A., Gettins P.G.W. quantitative dissection of the binding contributions of ligand lysines of the receptor-associated protein (RAP) to the low density lipoprotein receptor-related protein (LRP1) J Biol Chem. 2013;288:24081–24090. doi: 10.1074/jbc.M113.473728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Benchenane K., Berezowski V., Ali C., Fernandez-Monreal M., Lopez-Atalaya J.P., Brillault J. Tissue-type plasminogen activator crosses the intact blood-brain barrier by low-density lipoprotein receptor-related protein-mediated transcytosis. Circulation. 2005;111:2241–2249. doi: 10.1161/01.CIR.0000163542.48611.A2. [DOI] [PubMed] [Google Scholar]
- 112.Kawasaki T., Kaku S., Takenaka T., Yanagi K., Ohshima N. Thrombolytic activity of YM866, a novel modified tissue-type plasminogen activator, in a photochemically induced platelet-rich thrombosis model. J Cardiovasc Pharmacol. 1994;23:884. doi: 10.1097/00005344-199406000-00005. [DOI] [PubMed] [Google Scholar]
- 113.Bassel-Duby R., Jiang N.Y., Bittick T., Madison E., McGookey D., Orth K. Tyrosine 67 in the epidermal growth factor-like domain of tissue-type plasminogen activator is important for clearance by a specific hepatic receptor. J Biol Chem. 1992;267:9668–9677. [PubMed] [Google Scholar]
- 114.Goulay R., Naveau M., Gaberel T., Vivien D., Parcq J. Optimized tPA: a non-neurotoxic fibrinolytic agent for the drainage of intracerebral hemorrhages. J Cereb Blood Flow Metab. 2018;38:1180–1189. doi: 10.1177/0271678X17719180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Armstead W.M., Riley J., Yarovoi S., Higazi A.A.-R., Cines D.B. Tissue-type plasminogen activator-A(296-299) prevents impairment of cerebral autoregulation after stroke through lipoprotein-related receptor-dependent increase in cAMP and p38. Stroke. 2016;47:2096–2102. doi: 10.1161/STROKEAHA.116.012678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.López-Atalaya José P., Roussel Benoit D., Ali C., Maubert E., Petersen K.U., Berezowski V. Recombinant desmodus rotundus salivary plasminogen activator crosses the blood–brain barrier through a low-density lipoprotein receptor-related protein-dependent mechanism without exerting neurotoxic effects. Stroke. 2007;38:1036–1043. doi: 10.1161/01.STR.0000258100.04923.84. [DOI] [PubMed] [Google Scholar]
- 117.Madureira P.A., O’Connell P.A., Surette A.P., Miller V.A., Waisman D.M. The biochemistry and regulation of S100A10: A multifunctional plasminogen receptor involved in oncogenesis. J Biomed Biotechnol. 2012;2012:1–21. doi: 10.1155/2012/353687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee S.-B., Oh H.-K., Kim H.-K., Joe Y.A. Expression of the non-glycosylated kringle domain of tissue type plasminogen activator in Pichia and its anti-endothelial cell activity. Protein Expr Purif. 2006;50:1–8. doi: 10.1016/j.pep.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 119.Shim B.-S., Kang B.-H., Hong Y.-K., Kim H.-K., Lee I.-H., Lee S.-Y. The kringle domain of tissue-type plasminogen activator inhibits in vivo tumor growth. Biochem Biophys Res Commun. 2005;327:1155–1162. doi: 10.1016/j.bbrc.2004.12.126. [DOI] [PubMed] [Google Scholar]
- 120.Kim H.-K., Lee S.Y., Oh H.-K., Kang B.-H., Ku H.-J., Lee Y. Inhibition of endothelial cell proliferation by the recombinant kringle domain of tissue-type plasminogen activator. Biochem Biophys Res Commun. 2003;304:740–746. doi: 10.1016/s0006-291x(03)00656-9. [DOI] [PubMed] [Google Scholar]
- 121.Bharadwaj A., Bydoun M., Holloway R., Waisman D. Annexin A2 heterotetramer: structure and function. Int J Mol Sci. 2013;14:6259–6305. doi: 10.3390/ijms14036259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim J. Annexin II: a plasminogen-plasminogen activator co-receptor. Front Biosci. 2002;7:d341. doi: 10.2741/kim. [DOI] [PubMed] [Google Scholar]
- 123.Lin L., Wu C., Hu K. Tissue plasminogen activator activates NF-κB through a pathway involving annexin A2/CD11b and integrin-linked kinase. J Am Soc Nephrol JASN. 2012;23:1329–1338. doi: 10.1681/ASN.2011111123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kohnert U., Rudolph R., Verheijen J., Weeningverhoeff E., Stern A., Opitz U. Biochemical-properties of the kringle 2 and protease domains are maintained in the refolded T-Pa deletion variant Bm 06.022. Protein Eng. 1992;5:93–100. doi: 10.1093/protein/5.1.93. [DOI] [PubMed] [Google Scholar]
- 125.Thomas G., Thibodeaux H., Errett C., Badillo J., Keyt B., Refino C. a long-half-life and fibrin-specific form of tissue-plasminogen activator in rabbit models of embolic stroke and peripheral bleeding. Stroke. 1994;25:2072–2078. doi: 10.1161/01.str.25.10.2072. [DOI] [PubMed] [Google Scholar]
- 126.Eastman D., Wurm F.M., Van Reis R., Higgins D.L. A region of tissue plasminogen activator that affects plasminogen activation differentially with various fibrin(ogen)-related stimulators. Biochemistry. 1992;31:419–422. doi: 10.1021/bi00117a016. [DOI] [PubMed] [Google Scholar]
- 127.Malcolm A.D., Keltai M., Walsh M.J. ESPRIT: a European study of the prevention of reocclusion after initial thrombolysis with duteplase in acute myocardial infarction. Eur Heart J. 1996;17:1522–1531. doi: 10.1093/oxfordjournals.eurheartj.a014716. [DOI] [PubMed] [Google Scholar]
- 128.Investigators InTIME-II. Intravenous NPA for the treatment of infarcting myocardium early; inTIME-II, a double-blind comparison of single-bolus lanoteplase vs accelerated alteplase for the treatment of patients with acute myocardial infarction. Eur Heart J. 2000;21:2005–2013. doi: 10.1053/euhj.2000.2498. [DOI] [PubMed] [Google Scholar]
- 129.Hansen L., Blue Y., Barone K., Collen D., Larsen G.R. Functional effects of asparagine-linked oligosaccharide on natural and variant human tissue-type plasminogen activator. J Biol Chem. 1988;263:15713–15719. [PubMed] [Google Scholar]
- 130.Oikawa K., Watanabe T., Higuchi S. Comparison of drug disposition between wild-type and novel tissue-type plasminogen activator pamiteplase in rats. Drug Metab Dispos. 2000;28:1087–1093. [PubMed] [Google Scholar]
- 131.Flemmig M., Melzig M.F. Serine-proteases as plasminogen activators in terms of fibrinolysis. J Pharm Pharmacol. 2012;64:1025–1039. doi: 10.1111/j.2042-7158.2012.01457.x. [DOI] [PubMed] [Google Scholar]
- 132.Kazemali M., Majidzadeh-A K., Sardari S., Saadatirad A.H., Khalaj V., Zarei N. Design of a novel chimeric tissue plasminogen activator with favorable Vampire bat plasminogen activator properties. Enzyme Microb Technol. 2014;67:82–86. doi: 10.1016/j.enzmictec.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 133.Gurewich V. Experiences with pro-urokinase and potentiation of its fibrinolytic effect by urokinase and by tissue plasminogen activator. J Am Coll Cardiol. 1987;10:16B–21B. doi: 10.1016/s0735-1097(87)80423-0. [DOI] [PubMed] [Google Scholar]
- 134.Pannell R., Black J., Gurewich V. Complementary modes of action of tissue-type plasminogen activator and pro-urokinase by which their synergistic effect on clot lysis may be explained. J Clin Invest. 1988;81:853–859. doi: 10.1172/JCI113394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gurewich V. Why so little progress in therapeutic thrombolysis? The current state of the art and prospects for improvement. J Thromb Thrombolysis. 2015;40:480–487. doi: 10.1007/s11239-015-1217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pannell R., Li S., Gurewich V. Fibrin-specific and effective clot lysis requires both plasminogen activators and for them to be in a sequential rather than simultaneous combination. J Thromb Thrombolysis. 2017;44:210–215. doi: 10.1007/s11239-017-1514-0. [DOI] [PubMed] [Google Scholar]
- 137.Gladysz R., Adriaenssens Y., De Winter H., Joossens J., Lambeir A.-M., Augustyns K. Discovery and SAR of novel and selective inhibitors of urokinase plasminogen activator (uPA) with an imidazo[1,2-a]pyridine scaffold. J Med Chem. 2015;58:9238–9257. doi: 10.1021/acs.jmedchem.5b01171. [DOI] [PubMed] [Google Scholar]
- 138.Rabbani S.A., Ateeq B., Arakelian A., Valentino M.L., Shaw D.E., Dauffenbach L.M. An anti-urokinase plasminogen activator receptor antibody (ATN-658) blocks prostate cancer invasion, migration, growth, and experimental skeletal metastasis in vitro and in vivo. Neoplasia N Y N. 2010;12:778–788. doi: 10.1593/neo.10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xu X., Cai Y., Wei Y., Donate F., Juarez J., Parry G. Identification of a new epitope in uPAR as a target for the cancer therapeutic monoclonal antibody ATN-658, a structural homolog of the uPAR binding integrin CD11b (αM) PLoS One. 2014;9 doi: 10.1371/journal.pone.0085349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bifulco K., Longanesi-Cattani I., Gala M., Di Carluccio G., Masucci M.T., Pavone V. The soluble form of urokinase receptor promotes angiogenesis through its Ser88-Arg-Ser-Arg-Tyr92 chemotactic sequence: new vessel formation by SRSRY sequence of u-PAR. J Thromb Haemost. 2010;8:2789–2799. doi: 10.1111/j.1538-7836.2010.04075.x. [DOI] [PubMed] [Google Scholar]
- 141.Heissig B., Eiamboonsert S., Salama Y., Shimazu H., Dhahri D., Munakata S. Cancer therapy targeting the fibrinolytic system. Adv Drug Deliv Rev. 2016;99:172–179. doi: 10.1016/j.addr.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 142.Zhao Y., Lyons C.E., Jr., Xiao A., Templeton D.J., Sang Q.A., Brew K. Urokinase directly activates matrix metalloproteinases-9: a potential role in glioblastoma invasion. Biochem Biophys Res Commun. 2008;369:1215–1220. doi: 10.1016/j.bbrc.2008.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shin S.M., Cho K.S., Choi M.S., Lee S.H., Han S.-H., Kang Y.-S. Urokinase-type plasminogen activator induces BV-2 microglial cell migration through activation of matrix metalloproteinase-9. Neurochem Res. 2010;35:976–985. doi: 10.1007/s11064-010-0141-3. [DOI] [PubMed] [Google Scholar]
- 144.Carriero M.V., Franco P., Votta G., Longanesi-Cattani I., Vento M.T., Masucci M.T. Regulation of cell migration and invasion by specific modules of uPA: mechanistic insights and specific inhibitors. Curr Drug Targets. 2011;12:1761–1771. doi: 10.2174/138945011797635777. [DOI] [PubMed] [Google Scholar]
- 145.Ass K., Ahmad A., Azmi A.S., Sarkar S.H., Sarkar F.H. Evolving role of uPA/uPAR system in human cancers. Cancer Treat Rev. 2008;34:122–136. doi: 10.1016/j.ctrv.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 146.Lorenzi V.D., Ferraris G.M.S., Madsen J.B., Lupia M., Andreasen P.A., Sidenius N. Urokinase links plasminogen activation and cell adhesion by cleavage of the RGD motif in vitronectin. EMBO Rep. 2016:17:982. doi: 10.15252/embr.201541681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gandhari M., Arens N., Majety M., Dorn-Beineke A., Hildenbrand R.L. Urokinase-type plasminogen activator induces proliferation in breast cancer cells. Int J Oncol Rep. 2006;28:1463–1470. https://www.ncbi.nlm.nih.gov/pubmed/16685447 [PubMed] [Google Scholar]
- 148.Mekkawy A.H., Pourgholami M.H., Morris D.L. Involvement of urokinase-type plasminogen activator system in cancer: an overview: uPA system in cancer. Med Res Rev. 2014;34:918–956. doi: 10.1002/med.21308. [DOI] [PubMed] [Google Scholar]
- 149.Degen S.J., Rajput B., Reich E. The human tissue plasminogen activator gene. J Biol Chem. 1986;261:6972–6985. [PubMed] [Google Scholar]
- 150.Stephens R.W., Bokman A.M., Myohanen H.T., Reisberg T., Tapiovaara H., Pedersen N. Heparin binding to the urokinase kringle domain. Biochemistry. 1992;31:7572–7579. doi: 10.1021/bi00148a019. [DOI] [PubMed] [Google Scholar]
- 151.Franco P., Iaccarino C., Chiaradonna F., Brandazza A., Iavarone C., Mastronicola M.R. Phosphorylation of human pro-urokinase on ser 138/303 impairs its receptor-dependent ability to promote myelomonocytic adherence and motility. J Cell Biol. 1997;137:779–791. doi: 10.1083/jcb.137.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Buko A.M., Kentzer E.J., Petros A., Menon G., Zuiderweg E.R., Sarin V.K. Characterization of a posttranslational fucosylation in the growth factor domain of urinary plasminogen activator. Proc Natl Acad Sci. 1991;88:3992–3996. doi: 10.1073/pnas.88.9.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goochee C.F., Gramer M.J., Andersen D.C., Bahr J.B., Rasmussen J.R. The oligosaccharides of glycoproteins: bioprocess factors affecting oligosaccharide structure and their effect on glycoprotein properties. Bio/Technology. 1991;9:1347. doi: 10.1038/nbt1291-1347. [DOI] [PubMed] [Google Scholar]
- 154.Kasai S., Arimura H., Nishida M., Suyama T. Proteolytic cleavage of single-chain pro-urokinase induces conformational change which follows activation of the zymogen and reduction of its high affinity for fibrin. J Biol Chem. 1985;260:12377–12381. [PubMed] [Google Scholar]
- 155.Behrens M.A., Botkjaer K.A., Goswami S., Oliveira C.L.P., Jensen J.K., Schar C.R. Activation of the zymogen to urokinase-type plasminogen activator is associated with increased interdomain flexibility. J Mol Biol. 2011;411:417–429. doi: 10.1016/j.jmb.2011.05.026. [DOI] [PubMed] [Google Scholar]
- 156.Fleury V., Lijnen H.R., Anglés-Cano E. Mechanism of the enhanced intrinsic activity of single-chain urokinase-type plasminogen activator during ongoing fibrinolysis. J Biol Chem. 1993;268:18554–18559. [PubMed] [Google Scholar]
- 157.Kromann-Hansen T., Louise Lange E., Peter Sørensen H., Hassanzadeh-Ghassabeh G., Huang M., Jensen J.K. Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator. Sci Rep. 2017;7 doi: 10.1038/s41598-017-03457-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu Z., Kromann-Hansen T., Lund I.K., Hosseini M., Jensen K.J., Høyer-Hansen G. Interconversion of active and inactive conformations of urokinase-type plasminogen activator. Biochemistry. 2012;51:7804–7811. doi: 10.1021/bi3005957. [DOI] [PubMed] [Google Scholar]
- 159.Kromann-Hansen T., Lange E.L., Lund I.K., Høyer-Hansen G., Andreasen P.A., Komives E.A. Ligand binding modulates the structural dynamics and activity of urokinase-type plasminogen activator: a possible mechanism of plasminogen activation. PLoS One. 2018;13 doi: 10.1371/journal.pone.0192661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pannell R., Gurewich V. Pro-urokinase: a study of its stability in plasma and of a mechanism for its selective fibrinolytic effect. Blood. 1986;67:1215–1223. [PubMed] [Google Scholar]
- 161.Sun Z., Liu J.-N. Mutagenesis at Pro309 of single-chain urokinase-type plasminogen activator alters its catalytic properties. Proteins Struct Funct Bioinforma. 2005;61:870–877. doi: 10.1002/prot.20686. [DOI] [PubMed] [Google Scholar]
- 162.Blasi F., Vassalli J.D., Danø K. Urokinase-type plasminogen activator: proenzyme, receptor, and inhibitors. J Cell Biol. 1987;104:801–804. doi: 10.1083/jcb.104.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gurewich V., Pannell R. Inactivation of single-chain urokinase (pro-urokinase) by thrombin and thrombin-like enzymes: relevance of the findings to the interpretation of fibrin-binding experiments. Blood. 1987;69:769–772. [PubMed] [Google Scholar]
- 164.Lin Z., Jiang L., Yuan C., Jensen J.K., Zhang X., Luo Z. Structural basis for recognition of urokinase-type plasminogen activator by plasminogen activator inhibitor-1. J Biol Chem. 2011;286:7027–7032. doi: 10.1074/jbc.M110.204537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nienaber V., Wang J.Y., Davidson D., Henkin J. Re-engineering of human urokinase provides a system for structure-based drug design at high resolution and reveals a novel structural subsite. J Biol Chem. 2000;275:7239–7248. doi: 10.1074/jbc.275.10.7239. [DOI] [PubMed] [Google Scholar]
- 166.Katz B.A., Luong C., Ho J.D., Somoza J.R., Gjerstad E., Tang J. Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA) J Mol Biol. 2004;344:527–547. doi: 10.1016/j.jmb.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 167.Li C.Y., de Veer S.J., Law R.H.P., Whisstock J.C., Craik D.J., Swedberg J.E. Characterising the subsite specificity of urokinase-type plasminogen activator and tissue-type plasminogen activator using a sequence-defined peptide aldehyde library. Chem BioChem. 2019;20:46–50. doi: 10.1002/cbic.201800395. [DOI] [PubMed] [Google Scholar]
- 168.van der Kaaden M.E., Rijken D.C., van Berkel T.J.C., Kuiper J. Plasma clearance of urokinase-type plasminogen activator. Fibrinolysis Proteolysis. 1998;12:251–258. [Google Scholar]
- 169.Croucher D., Saunders D.N., Ranson M. The urokinase/PAI-2 complex - A new high affinity ligand for the endocytosis receptor low density lipoprotein receptor-related protein. J Biol Chem. 2006;281:10206–10213. doi: 10.1074/jbc.M513645200. [DOI] [PubMed] [Google Scholar]
- 170.Kounnas M.Z., Henkin J., Argraves W.S., Strickland D.K. Low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor mediates cellular uptake of pro-urokinase. J Biol Chem. 1993;268:21862–21867. [PubMed] [Google Scholar]
- 171.Nykjaer A., Kjøller L., Cohen R.L., Lawrence D.A., Garni-Wagner B.A., Todd R.F. Regions involved in binding of urokinase-type-1 inhibitor complex and pro-urokinase to the endocytic alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein. evidence that the urokinase receptor protects pro-urokinase against binding to the endocytic receptor. J Biol Chem. 1994;269:25668–25676. [PubMed] [Google Scholar]
- 172.Skeldal S., Larsen J.V., Pedersen K.E., Petersen H.H., Egelund R., Christensen A. Binding areas of urokinase-type plasminogen activator–plasminogen activator inhibitor-1 complex for endocytosis receptors of the low-density lipoprotein receptor family, determined by site-directed mutagenesis. FEBS J. 2006;273:5143–5159. doi: 10.1111/j.1742-4658.2006.05511.x. [DOI] [PubMed] [Google Scholar]
- 173.Ellis V., Whawell S.A., Werner F., Deadman J.J. Assembly of urokinase receptor-mediated plasminogen activation complexes involves direct, non-active-site interactions between urokinase and plasminogen. Biochemistry. 1999;38:651–659. doi: 10.1021/bi981714d. [DOI] [PubMed] [Google Scholar]
- 174.Huai Q., Mazar A.P., Kuo A., Parry G.C., Shaw D.E., Callahan J. Structure of human urokinase plasminogen activator in complex with its receptor. Science. 2006;311:656–659. doi: 10.1126/science.1121143. [DOI] [PubMed] [Google Scholar]
- 175.Ploug M., Gårdsvoll H., Jørgensen T.J.D., Hansen L.L., Danø K. Structural analysis of the interaction between urokinase-type plasminogen activator and its receptor: a potential target for anti-invasive cancer therapy. Biochem Soc Trans. 2002;30:177–183. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 176.Appella E., Robinson E.A., Ullrich S.J., Stoppelli M.P., Corti A., Cassani C. In: The receptor-binding sequence of urokinase. Walsh K.A., editor. Humana Press; Totowa, NJ: 1986. pp. 551–554. Methods Protein Seq. Anal.. 1987. [PubMed] [Google Scholar]
- 177.Degryse B., Fernandez-Recio J., Citro V., Blasi F., Cubellis M.V. In silico docking of urokinase plasminogen activator and integrins. BMC Bioinformatics. 2008;9:S8. doi: 10.1186/1471-2105-9-S2-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Franco P., Carotenuto A., Marcozzi C., Votta G., Sarno C., Iaccarino I. opposite modulation of cell migration by distinct subregions of urokinase connecting peptide. Chembiochem. 2013;14:882–889. doi: 10.1002/cbic.201200774. [DOI] [PubMed] [Google Scholar]
- 179.Novokhatny V., Medved L., Lijnen H.R., Ingham K. Tissue-type plasminogen activator (tPA) interacts with urokinase-type plasminogen activator (uPA) via tPA’s lysine binding site: an explanation of the poor fibrin affinity of recombinant tPA/uPA chimeric molecules. J Biol Chem. 1995;270:8680–8685. doi: 10.1074/jbc.270.15.8680. [DOI] [PubMed] [Google Scholar]
- 180.Sun Z., Jiang Y., Ma Z., Wu H., Liu B.-F., Xu Y. Identification of a flexible loop region (297–313) of urokinase-type plasminogen activator, which helps determine its catalytic activity. J Biol Chem. 1997;272:23818–23823. doi: 10.1074/jbc.272.38.23818. [DOI] [PubMed] [Google Scholar]
- 181.Gurewich V., Pannell R., Simmons-Byrd A., Sarmientos P., Liu J.-N., Badylak S.F. Thrombolysis vs. bleeding from hemostatic sites by a prourokinase mutant compared with tissue plasminogen activator. J Thromb Haemost. 2006;4:1559–1565. doi: 10.1111/j.1538-7836.2006.01993.x. [DOI] [PubMed] [Google Scholar]
- 182.Tillett W.S., Garner R.L. The fibrinolytic activity of hemolytic streptococci. J Exp Med. 1933;58:485–502. doi: 10.1084/jem.58.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Broder C.C., Lottenberg R., von Mering G.O., Johnston K.H., Boyle M.D. Isolation of a prokaryotic plasmin receptor. relationship to a plasminogen activator produced by the same micro-organism. J Biol Chem. 1991;266:4922–4928. [PubMed] [Google Scholar]
- 184.Sun H., Ringdahl U., Homeister J.W., Fay W.P., Engleberg N.C., Yang A.Y. Plasminogen is a critical host pathogenicity factor for group a streptococcal infection. Science. 2004;305:1283–1286. doi: 10.1126/science.1101245. [DOI] [PubMed] [Google Scholar]
- 185.Huish S., Thelwell C., Longstaff C. Activity regulation by fibrinogen and fibrin of streptokinase from streptococcus pyogenes. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Reed G.L., Houng A.K., Liu L., Parhami-Seren B., Matsueda L.H., Wang S.G. A catalytic switch and the conversion of streptokinase to a fibrin-targeted plasminogen activator. Proc Natl Acad Sci U S A. 1999;96:8879–8883. doi: 10.1073/pnas.96.16.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Banerjee A., Chisti Y., Banerjee U.C. Streptokinase - a clinically useful thrombolytic agent. Biotechnol Adv. 2004;22:287–307. doi: 10.1016/j.biotechadv.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 188.Lee H.S. How safe is the readministration of streptokinase? Drug Saf. 1995;13:76–80. doi: 10.2165/00002018-199513020-00002. [DOI] [PubMed] [Google Scholar]
- 189.Jennings K. Antibodies to streptokinase. BMJ. 1996;312:393–394. doi: 10.1136/bmj.312.7028.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Malke H., Roe B., Ferretti J.J. Nucleotide sequence of the streptokinase gene from Streptococcus equisimilis H46A. Gene. 1985;34:357–362. doi: 10.1016/0378-1119(85)90145-3. [DOI] [PubMed] [Google Scholar]
- 191.Renzo E.C.D., Siiteri P.K., Hutchings B.L., Bell P.H. Preparation and certain properties of highly purified streptokinase. J Biol Chem. 1967;242:533–542. [PubMed] [Google Scholar]
- 192.Teuten A.J., Broadhurst R.W., Smith R.A.G., Dobson C.M. Characterization of structural and folding properties of streptokinase by n.m.r. spectroscopy. Biochem J. 1993;290:313–319. doi: 10.1042/bj2900313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wang X., Lin X., Loy J.A., Tang J., Zhang X.C. Crystal structure of the catalytic domain of human plasmin complexed with streptokinase. Science. 1998;281:1662–1665. doi: 10.1126/science.281.5383.1662. [DOI] [PubMed] [Google Scholar]
- 194.Sazonova I.Y., Robinson B.R., Gladysheva I.P., Castellino F.J., Reed G.L. α Domain deletion converts streptokinase into a fibrin-dependent plasminogen activator through mechanisms akin to staphylokinase and tissue plasminogen activator. J Biol Chem. 2004;279:24994–25001. doi: 10.1074/jbc.M400253200. [DOI] [PubMed] [Google Scholar]
- 195.Wang S.G., Reed G.L., Hedstrom L. Deletion of Ile1 changes the mechanism of streptokinase: evidence for the molecular sexuality hypothesis. Biochemistry. 1999;38:5232–5240. doi: 10.1021/bi981915h. [DOI] [PubMed] [Google Scholar]
- 196.Wang S.G., Reed G.L., Hedstrom L. Zymogen activation in the streptokinase-plasminogen complex - Ile1 is required for the formation of a functional active site. Eur J Biochem. 2000;267:3994–4001. doi: 10.1046/j.1432-1327.2000.01434.x. [DOI] [PubMed] [Google Scholar]
- 197.Loy J.A., Lin X., Schenone M., Castellino F.J., Zhang X.C., Tang J. Domain interactions between streptokinase and human plasminogen †. Biochemistry. 2001;40:14686–14695. doi: 10.1021/bi011309d. [DOI] [PubMed] [Google Scholar]
- 198.Liu L., Sazonova I.Y., Turner R.B., Chowdhry S.A., Tsai J., Houng A.K. Leucine 42 in the fibronectin motif of streptokinase plays a critical role in fibrin-independent plasminogen activation. J Biol Chem. 2000;275:37686–37691. doi: 10.1074/jbc.M003963200. [DOI] [PubMed] [Google Scholar]
- 199.Boxrud P.D., Verhamme I.M.A., Fay W.P., Bock P.E. Streptokinase triggers conformational activation of plasminogen through specific interactions of the amino-terminal sequence and stabilizes the active zymogen conformation. J Biol Chem. 2001;276:26084–26089. doi: 10.1074/jbc.M101966200. [DOI] [PubMed] [Google Scholar]
- 200.Mundada L.V., Prorok M., DeFord M.E., Figuera M., Castellino F.J., Fay W.P. Structure-function analysis of the streptokinase amino terminus (residues 1-59) J Biol Chem. 2003;278:24421–24427. doi: 10.1074/jbc.M301825200. [DOI] [PubMed] [Google Scholar]
- 201.Young K.-C., Shi G.-Y., Wu D.-H., Chang L.-C., Chang B.-I., Ou C.-P. Plasminogen activation by streptokinase via a unique mechanism. J Biol Chem. 1998;273:3110–3116. doi: 10.1074/jbc.273.5.3110. [DOI] [PubMed] [Google Scholar]
- 202.Wakeham N., Terzyan S., Zhai P.Z., Loy J.A., Tang J., Zhang X.C. Effects of deletion of streptokinase residues 48-59 on plasminogen activation. Protein Eng. 2002;15:753–761. doi: 10.1093/protein/15.9.753. [DOI] [PubMed] [Google Scholar]
- 203.Kim D.M., Lee S.J., Kim I.C., Kim S.T., Byun S.M. Asp41-His48 region of streptokinase is important in binding to a substrate plasminogen. Thromb Res. 2000;99:93–98. doi: 10.1016/s0049-3848(00)00225-5. [DOI] [PubMed] [Google Scholar]
- 204.Parrado J., Conejero-Lara F., Smith R.A., Marshall J.M., Ponting C.P., Dobson C.M. The domain organization of streptokinase: nuclear magnetic resonance, circular dichroism, and functional characterization of proteolytic fragments. Protein Sci Publ Protein Soc. 1996;5:693–704. doi: 10.1002/pro.5560050414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Rodríguez P., Fuentes P., Barro M., Alvarez J.G., Muñoz E., Collen D. Structural domains of streptokinase involved in the interaction with plasminogen. Eur J Biochem. 1995;229:83–90. doi: 10.1111/j.1432-1033.1995.tb20441.x. [DOI] [PubMed] [Google Scholar]
- 206.Conejero-Lara F., Parrado J., Azuaga A.I., Dobson C.M., Ponting C.P. Analysis of the interactions between streptokinase domains and human plasminogen. Protein Sci Publ Protein Soc. 1998;7:2190–2199. doi: 10.1002/pro.5560071017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chaudhary A., Vasudha S., Rajagopal K., Komath S.S., Garg N., Yadav M. Function of the central domain of streptokinase in substrate plasminogen docking and processing revealed by site-directed mutagenesis. Protein Sci. 1999;8:2791–2805. doi: 10.1110/ps.8.12.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lin L.F., Oeun S., Houng A., Reed G.L. Mutation of lysines in a plasminogen binding region of streptokinase identifies residues important for generating a functional activator complex. Biochemistry. 1996;35:16879–16885. doi: 10.1021/bi961531w. [DOI] [PubMed] [Google Scholar]
- 209.Aneja R., Datt M., Singh B., Kumar S., Sahni G. Identification of a new exosite involved in catalytic turnover by the streptokinase-plasmin activator complex during human plasminogen activation. J Biol Chem. 2009;284:32642–32650. doi: 10.1074/jbc.M109.046573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Dhar J., Pande A.H., Sundram V., Nanda J.S., Mande S.C., Sahni G. Involvement of a nine-residue loop of streptokinase in the generation of macromolecular substrate specificity by the activator complex through interaction with substrate kringle domains. J Biol Chem. 2002;277:13257–13267. doi: 10.1074/jbc.M108422200. [DOI] [PubMed] [Google Scholar]
- 211.Tharp A.C., Laha M., Panizzi P., Thompson M.W., Fuentes-Prior P., Bock P.E. Plasminogen substrate recognition by the streptokinase-plasminogen catalytic complex is facilitated by Arg253, Lys256, and Lys257 in the streptokinase β-domain and kringle 5 of the substrate. J Biol Chem. 2009;284:19511–19521. doi: 10.1074/jbc.M109.005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wu D.H., Shi G.Y., Chuang W.J., Hsu J.M., Young K.C., Chang C.W. Coiled coil region of streptokinase gamma-domain is essential for plasminogen activation. J Biol Chem. 2001;276:15025–15033. doi: 10.1074/jbc.M005935200. [DOI] [PubMed] [Google Scholar]
- 213.Yadav S., Aneja R., Kumar P., Datt M., Sinha S., Sahni G. Identification through combinatorial random and rational mutagenesis of a substrate-interacting exosite in the γ domain of streptokinase. J Biol Chem. 2011;286:6458–6469. doi: 10.1074/jbc.M110.152355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Panizzi P., Boxrud P.D., Verhamme I.M., Bock P.E. Binding of the COOH-terminal lysine residue of streptokinase to plasmin(ogen) kringles enhances formation of the streptokinase center dot plasmin(ogen) catalytic complexes. J Biol Chem. 2006;281:26774–26778. doi: 10.1074/jbc.C600171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Aneja R., Datt M., Yadav S., Sahni G. Multiple exosites distributed across the three domains of streptokinase co-operate to generate high catalytic rates in the streptokinase–plasmin activator complex. Biochemistry. 2013;52:8957–8968. doi: 10.1021/bi400142s. [DOI] [PubMed] [Google Scholar]
- 216.Sundram V., Nanda J.S., Rajagopal K., Dhar J., Chaudhary A., Sahni G. Domain truncation studies reveal that the streptokinase-plasmin activator complex utilizes long range protein-protein interactions with macromolecular substrate to maximize catalytic turnover. J Biol Chem. 2003;278:30569–30577. doi: 10.1074/jbc.M303799200. [DOI] [PubMed] [Google Scholar]
- 217.Reddy K.N.N., Markus G. Mechanism of activation of human plasminogen by streptokinase. Presence of active center in streptokinase-plasminogen complex. J Biol Chem. 1972;247:1683–1691. [PubMed] [Google Scholar]
- 218.McClintock D.K., Bell P.H. The mechanism of activation of human plasminogen by streptokinase. Biochem Biophys Res Commun. 1971;43:694–702. doi: 10.1016/0006-291x(71)90670-x. [DOI] [PubMed] [Google Scholar]
- 219.Nolan M., Bouldin S.D., Bock P.E. Full time course kinetics of the streptokinase-plasminogen activation pathway. J Biol Chem. 2013;288:29482–29493. doi: 10.1074/jbc.M113.477935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lin L.F., Houng A.Y., Reed G.L. Epsilon amino caproic acid inhibits streptokinase-plasminogen activator complex formation and substrate binding through kringle-dependent mechanisms. Biochemistry. 2000;39:4740–4745. doi: 10.1021/bi992028x. [DOI] [PubMed] [Google Scholar]
- 221.Boxrud P.D., Bock P.E. Streptokinase binds preferentially to the extended conformation of plasminogen through lysine binding site and catalytic domain interactions. Biochemistry. 2000;39:13974–13981. doi: 10.1021/bi000594i. [DOI] [PubMed] [Google Scholar]
- 222.Verhamme I.M., Bock P.E. Rapid binding of plasminogen to streptokinase in a catalytic complex reveals a three-step mechanism. J Biol Chem. 2014;289:28006–28018. doi: 10.1074/jbc.M114.589077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Verhamme I.M., Bock P.E. Rapid-reaction kinetic characterization of the pathway of streptokinase-plasmin catalytic complex formation. J Biol Chem. 2008;283:26137–26147. doi: 10.1074/jbc.M804038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yadav S., Datt M., Singh B., Sahni G. Role of the 88-97 loop in plasminogen activation by streptokinase probed through site-specific mutagenesis. Biochim Biophys Acta-Proteins Proteomics. 1784;2008:1310–1318. doi: 10.1016/j.bbapap.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 225.Boxrud P.D., Bock P.E. Coupling of conformational and proteolytic activation in the kinetic mechanism of plasminogen activation by streptokinase. J Biol Chem. 2004;279:36642–36649. doi: 10.1074/jbc.M405265200. [DOI] [PubMed] [Google Scholar]
- 226.Collen D., Hoef B.V., Schlott B., Hartmann M., Gührs K.-H., Lijnen H.R. Mechanisms of activation of mammalian plasma fibrinolytic systems with streptokinase and with recombinant staphylokinase. Eur J Biochem. 1993;216:307–314. doi: 10.1111/j.1432-1033.1993.tb18147.x. [DOI] [PubMed] [Google Scholar]
- 227.Lijnen H.R., Van Hoef B., De Cock F., Okada K., Ueshima S., Matsuo O. On the mechanism of fibrin-specific plasminogen activation by staphylokinase. J Biol Chem. 1991;266:11826–11832. [PubMed] [Google Scholar]
- 228.Silence K., Collen D., Lijnen H.R. Regulation by alpha 2-antiplasmin and fibrin of the activation of plasminogen with recombinant staphylokinase in plasma. Blood. 1993;82:1175–11783. [PubMed] [Google Scholar]
- 229.Sazonova I.Y., McNamee R.A., Houng A.K., King S.M., Hedstrom L., Reed G.L. Reprogrammed streptokinases develop fibrin-targeting and dissolve blood clots with more potency than tissue plasminogen activator. J Thromb Haemost. 2009;7:1321–1328. doi: 10.1111/j.1538-7836.2009.03491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Cook S.M., Skora A., Walker M.J., Sanderson-Smith M.L., McArthur J.D. Site-restricted plasminogen activation mediated by group A streptococcal streptokinase variants. Biochem J. 2014;458:23–31. doi: 10.1042/BJ20131305. [DOI] [PubMed] [Google Scholar]
- 231.Taheri M.N., Behzad-Behbahani A., Rafiei Dehbidi G., Salehi S., Sharifzadeh S. Engineering, expression and purification of a chimeric fibrin-specific streptokinase. Protein Expr Purif. 2016;128:14–21. doi: 10.1016/j.pep.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 232.Zhang Y., Gladysheva I.P., Houng A.K., Reed G.L. Streptococcus uberis plasminogen activator (SUPA) activates human plasminogen through novel species-specific and fibrin-targeted mechanisms. J Biol Chem. 2012;287:19171–19176. doi: 10.1074/jbc.M112.359315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Boxrud P.D., Fay W.P., Bock P.E. Streptokinase binds to human plasmin with high affinity, perturbs the plasmin active site, and induces expression of a substrate recognition exosite for plasminogen. J Biol Chem. 2000;275:14579–14589. doi: 10.1074/jbc.275.19.14579. [DOI] [PubMed] [Google Scholar]
- 234.Shi G.Y., Chang B.I., Chen S.M., Wu D.H., Wu H.L. Function of streptokinase fragments in plasminogen activation. Biochem J. 1994;304:235–241. doi: 10.1042/bj3040235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Adivitiya, Babbal, Mohanty S., Khasa Y.P. Engineering of deglycosylated and plasmin resistant variants of recombinant streptokinase in Pichia pastoris. Appl Microbiol Biotechnol. 2018;102:10561–10577. doi: 10.1007/s00253-018-9402-x. [DOI] [PubMed] [Google Scholar]
- 236.Wu X.-C., Ye R., Duan Y., Wong S.-L. Engineering of plasmin-resistant forms of streptokinase and their production in bacillus subtilis: streptokinase with longer functional half-life. Appl Environ Microbiol. 1998;64:824–829. doi: 10.1128/aem.64.3.824-829.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Shi G.-Y., Chang B.-I., Su S.-W., Young K.-C., Wu D.-H., Chang L.-C. Preparation of a novel streptokinase mutant with improved stability. Thromb Haemost. 1998;79:992–997. [PubMed] [Google Scholar]
- 238.Wong S.L., Ye R., Nathoo S. Engineering and production of streptokinase in a Bacillus subtilis expression-secretion system. Appl Environ Microbiol. 1994;60:517–523. doi: 10.1128/aem.60.2.517-523.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sawhney P., Katare K., Sahni G. PEGylation of truncated streptokinase leads to formulation of a useful drug with ameliorated attributes. Plos One. 2016;11 doi: 10.1371/journal.pone.0155831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sawhney P., Kumar S., Maheshwari N., Guleria S.S., Dhar N., Kashyap R. Site-Specific Thiol-mediated PEGylation of streptokinase leads to improved properties with clinical potential. Curr Pharm Des. 2016;22:5868–5878. doi: 10.2174/1381612822666160204120547. [DOI] [PubMed] [Google Scholar]
- 241.Rajagopalan S., Gonias S.L., Pizzo S.V. A nonantigenic covalent streptokinase-polyethylene glycol complex with plasminogen activator function. J Clin Invest. 1985;75:413–419. doi: 10.1172/JCI111715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Koide A., Suzuki S., Kobayashi S. Preparation of polyethylene glycol-modified streptokinase with disappearance of binding ability towards anti-serum and retention of activity. FEBS Lett. 1982;143:73–76. doi: 10.1016/0014-5793(82)80276-7. [DOI] [PubMed] [Google Scholar]
- 243.Pratap J., Rajamohan G., Dikshit K.L. Characteristics of glycosylated streptokinase secreted from Pichia pastoris: enhanced resistance of SK to proteolysis by glycosylation. Appl Microbiol Biotechnol. 2000;53:469–475. doi: 10.1007/s002530051643. [DOI] [PubMed] [Google Scholar]
- 244.Suthakaran P., Balasubramanian J., Ravichandran M., Murugan V., Ramya L.N., Pulicherla K.K. Studies on lipidification of streptokinase: a novel strategy to enhance the stability and activity. Am J Ther. 2014;21:343. doi: 10.1097/MJT.0000000000000029. [DOI] [PubMed] [Google Scholar]
- 245.Smith R.A.G., Dupe R.J., English P.D., Green J. Fibrinolysis with acyl-enzymes: a new approach to thrombolytic therapy. Nature. 1981;290:505–508. doi: 10.1038/290505a0. [DOI] [PubMed] [Google Scholar]
- 246.Monk J.P., Heel R.C. Anisoylated plasminogen streptokinase activator complex (APSAC). A review of its mechanism of action, clinical pharmacology and therapeutic use in acute myocardial infarction. Drugs. 1987;34:25–49. doi: 10.2165/00003495-198734010-00002. [DOI] [PubMed] [Google Scholar]
- 247.Anderson J.L. Development and evaluation of anisoylated plasminogen streptokinase activator complex (APSAC) as a second generation thrombolytic agent. J Am Coll Cardiol. 1987;10:22B–27B. doi: 10.1016/s0735-1097(87)80424-2. [DOI] [PubMed] [Google Scholar]
- 248.Lack C.H. Staphylokinase : an activator of plasma protease. Nature. 1948;161:559–560. doi: 10.1038/161559b0. [DOI] [PubMed] [Google Scholar]
- 249.Sako T., Sawaki S., Sakurai T., Ito S., Yoshizawa Y., Kondo I. Cloning and expression of the staphylokinase gene of Staphylococcus aureus in Escherichia coli. Mol Gen Genet. 1983;190:271–277. doi: 10.1007/BF00330650. [DOI] [PubMed] [Google Scholar]
- 250.Collen D., De Cock F., Vanlinthout I., Declerck P.J., Lijnen H.R., Stassen J.M. Comparative thrombolytic and immunogenic properties of staphylokinase and streptokinase. Fibrinolysis. 1992;6:232–242. [Google Scholar]
- 251.Collen D., Lijnen H.R. Staphylokinase, a fibrin-specific plasminogen activator with therapeutic potential? Blood. 1994;84:680–686. [PubMed] [Google Scholar]
- 252.Collen D. Staphylokinase: a potent, uniquely fibrin-selective thrombolytic agent. Nat Med. 1998;4:279–284. doi: 10.1038/nm0398-279. [DOI] [PubMed] [Google Scholar]
- 253.Matsuo O., Okada K., Fukao H., Tomioka Y., Ueshima S., Watanuki M. Thrombolytic properties of staphylokinase. Blood. 1990;76:925–929. [PubMed] [Google Scholar]
- 254.Behnke D., Gerlach D. Cloning and expression in Escherichia coli, Bacillus subtilis, and Streptococcus sanguis of a gene for staphylokinase — a bacterial plasminogen activator. Mol Gen Genet MGG. 1987;210:528–534. doi: 10.1007/BF00327208. [DOI] [PubMed] [Google Scholar]
- 255.Collen D., Zhao Z.A., Holvoet P., Marynen P. Primary structure and gene structure of staphylokinase. Fibrinolysis. 1992;6:226–231. [Google Scholar]
- 256.Rabijns A., Bondt H.L.D., Ranter C.D. Three-dimensional structure of staphylokinase, a plasminogen activator with therapeutic potential. Nat Struct Biol. 1997;4:357–360. doi: 10.1038/nsb0597-357. [DOI] [PubMed] [Google Scholar]
- 257.Parry M.A., Fernandez-Catalan C., Bergner A., Huber R., Hopfner K.P., Schlott B. The ternary microplasmin-staphylokinase-microplasmin complex is a proteinase-cofactor-substrate complex in action. Nat Struct Biol. 1998;5:917–923. doi: 10.1038/2359. [DOI] [PubMed] [Google Scholar]
- 258.Ohlenschläger O., Ramachandran R., Flemming J., Gührs K.-H., Schlott B., Brown L.R. NMR secondary structure of the plasminogen activator protein staphylokinase. J Biomol NMR. 1997;9:273–286. doi: 10.1023/a:1018678925512. [DOI] [PubMed] [Google Scholar]
- 259.Kowalska-Loth B., Zakrzewski K. The activation by staphylokinase of human plasminogen. Acta Biochim Pol. 1975;22:327–339. [PubMed] [Google Scholar]
- 260.Grella D.K., Castellino F.J. Activation of human plasminogen by staphylokinase. direct evidence that preformed plasmin is necessary for activation to occur. Blood. 1997;89:1585–1589. [PubMed] [Google Scholar]
- 261.Collen D., Schlott B., Engelborghs Y., Hoef B.V., Hartmann M., Lijnen H.R. On the mechanism of the activation of human plasminogen by recombinant staphylokinase. J Biol Chem. 1993;268:8284–8289. [PubMed] [Google Scholar]
- 262.Shibata H., Nagaoka M., Sakai M., Sawada H., Watanabe T., Yokokura T. Kinetic studies on the plasminogen activation by the staphylokinase-plasmin complex. J Biochem (Tokyo) 1994;115:738–742. doi: 10.1093/oxfordjournals.jbchem.a124404. [DOI] [PubMed] [Google Scholar]
- 263.Schlott B., Hartmann M., Gührs K.-H., Birch-Hirschfeld E., Gase A., Vettermann S. Functional properties of recombinant staphylokinase variants obtained by site-specific mutagenesis of methionine-26. Biochim Biophys Acta BBA Protein Struct Mol Enzymol. 1994;1204:235–242. doi: 10.1016/0167-4838(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 264.Dahiya M., Singh S., Rajamohan G., Sethi D., Ashish Dikshit K.L. Intermolecular interactions in staphylokinase-plasmin(ogen) bimolecular complex: function of His43 and Tyr44. FEBS Lett. 2011;585:1814–1820. doi: 10.1016/j.febslet.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 265.Singh S., Ashish Dikshit K.L. Pro(42) and Val(45) of staphylokinase modulate intermolecular interactions of His(43)-Tyr(44) pair and specificity of staphylokinase-plasmin activator complex. FEBS Lett. 2012;586:653–658. doi: 10.1016/j.febslet.2012.01.046. [DOI] [PubMed] [Google Scholar]
- 266.Silence K., Hartmann M., Gührs K.-H., Gase A., Schlott B., Collen D. Structure-function relationships in staphylokinase as revealed by “clustered charge to alanine” mutagenesis. J Biol Chem. 1995;270:27192–27198. doi: 10.1074/jbc.270.45.27192. [DOI] [PubMed] [Google Scholar]
- 267.Jespers L., Lijnen H.R., Vanwetswinkel S., Van Hoef B., Brepoels K., Collen D. Guiding a docking mode by phage display: selection of correlated mutations at the staphylokinase-plasmin interface11edited by A. R Fersht J Mol Biol. 1999;290:471–479. doi: 10.1006/jmbi.1999.2887. [DOI] [PubMed] [Google Scholar]
- 268.Schlott B., Gührs K.-H., Hartmann M., Röcker A., Collen D. NH2-terminal structural motifs in staphylokinase required for plasminogen activation. J Biol Chem. 1998;273:22346–22350. doi: 10.1074/jbc.273.35.22346. [DOI] [PubMed] [Google Scholar]
- 269.Rajamohan G., Dikshit K.L. Role of the N-terminal region of staphylokinase (SAK): evidence for the participation of the N-terminal region of SAK in the enzyme–substrate complex formation. FEBS Lett. 2000;474:151–158. doi: 10.1016/s0014-5793(00)01578-7. [DOI] [PubMed] [Google Scholar]
- 270.Ohlenschläger O., Ramachandran R., Gührs K.-H., Schlott B., Brown L.R. Nuclear magnetic resonance solution structure of the plasminogen-activator protein staphylokinase. Biochemistry. 1998;37:10635–10642. doi: 10.1021/bi980673i. [DOI] [PubMed] [Google Scholar]
- 271.Trieu T., Behnke D., Gerlach D., Tang J. [9] Activation of human plasminogen by recombinant staphylokinase. Methods Enzymol. 1993;223:156–167. doi: 10.1016/0076-6879(93)23043-m. Academic Press. [DOI] [PubMed] [Google Scholar]
- 272.Lijnen H.R., Van Hoef B., Vandenbossche L., Collen D. Biochemical properties of natural and recombinant staphylokinase. Fibrinolysis. 1992;6:214–225. [Google Scholar]
- 273.Ueshima S., Silence K., Collen D., Lijnen H.R. Molecular conversions of recombinant staphylokinase during plasminogen activation in purified systems and in human plasma. Thromb Haemost. 1993;70:495–499. [PubMed] [Google Scholar]
- 274.Collen D., Silence K., Demarsin E., De Mol M., Lijnen H.R. Isolation and characterisation of natural and recombinant staphylokinase. Fibrinolysis. 1992;6:203–213. [Google Scholar]
- 275.Schlott B., Gührs K.-H., Hartmann M., Röcker A., Collen D. Staphylokinase requires NH2-terminal proteolysis for plasminogen activation. J Biol Chem. 1997;272:6067–6072. doi: 10.1074/jbc.272.9.6067. [DOI] [PubMed] [Google Scholar]
- 276.Sakai M., Watanuki M., Matsuo O. Mechanism of fibrin-specific fibrinolysis by staphylokinase: participation of α2-plasmin inhibitor. Biochem Biophys Res Commun. 1989;162:830–837. doi: 10.1016/0006-291x(89)92385-1. [DOI] [PubMed] [Google Scholar]
- 277.Lijnen H.R., Van Hoef B., Matsuo Osamu, Collen D. On the molecular interactions between plasminogen-staphylokinase, α2-antiplasmin and fibrin. Biochim Biophys Acta BBA Protein Struct Mol Enzymol. 1992;1118:144–148. doi: 10.1016/0167-4838(92)90142-z. [DOI] [PubMed] [Google Scholar]
- 278.Okada K., Ueshima S., Takaishi T., Yuasa H., Fukao H., Matsuo O. Effects of fibrin and α2-antiplasmin on plasminogen activation by staphylokinase. Am J Hematol. 1996;53:151–157. doi: 10.1002/(SICI)1096-8652(199611)53:3<151::AID-AJH1>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 279.Lijnen H.R., Hoef B.V., Collen D. Interaction of staphylokinase with different molecular forms of plasminogen. Eur J Biochem. 1993;211:91–97. doi: 10.1111/j.1432-1033.1993.tb19873.x. [DOI] [PubMed] [Google Scholar]
- 280.Sakharov D.V., Lijnen H.R., Rijken D.C. Interactions between staphylokinase, plasmin(ogen), and fibrin. staphylokinase discriminates between free plasminogen and plasminogen bound to partially degraded fibrin. J Biol Chem. 1996;271:27912–27918. doi: 10.1074/jbc.271.44.27912. [DOI] [PubMed] [Google Scholar]
- 281.Silence K., Collen D., Lijnen H.R. Interaction between staphylokinase, plasmin(ogen), and alpha 2-antiplasmin. Recycling of staphylokinase after neutralization of the plasmin-staphylokinase complex by alpha 2-antiplasmin. J Biol Chem. 1993;268:9811–9816. [PubMed] [Google Scholar]
- 282.Collen D. Fibrin-selective thrombolytic therapy for acute myocardial infarction. Circulation. 1996;93:857–865. doi: 10.1161/01.cir.93.5.857. [DOI] [PubMed] [Google Scholar]
- 283.Collen D. Engineered staphylokinase variants with reduced immunogenicity. Fibrinolysis Proteolysis. 1998;12:59–65. [Google Scholar]
- 284.Jespers L., Jenné S., Lasters I., Collen D. Epitope mapping by negative selection of randomized antigen libraries displayed on filamentous phage11Edited by J. Karn J Mol Biol. 1997;269:704–718. doi: 10.1006/jmbi.1997.1077. [DOI] [PubMed] [Google Scholar]
- 285.Jenné S., Brepoels K., Collen D., Jespers L. High resolution mapping of the B cell epitopes of staphylokinase in humans using negative selection of a phage-displayed antigen library. J Immunol. 1998;161:3161–3168. [PubMed] [Google Scholar]
- 286.Laroche Y., Heymans S., Capaert S., Cock F.D., Demarsin E., Collen D. Recombinant staphylokinase variants with reduced antigenicity due to elimination of B-lymphocyte epitopes. Blood. 2000;96:1425–1432. [PubMed] [Google Scholar]
- 287.He J., Xu R., Chen X., Jia K., Zhou X., Zhu K. Simultaneous elimination of T- and B-cell epitope by structure-based mutagenesis of single Glu80 residue within recombinant staphylokinase. Acta Biochim Biophys Sin. 2010;42:209–215. doi: 10.1093/abbs/gmq002. [DOI] [PubMed] [Google Scholar]
- 288.Su H.-B., Zhang Y.-G., He J.-T., Mo W., Zhang Y.-L., Tao X.-M. Construction and characterization of novel staphylokinase variants with antiplatelet aggregation activity and reduced immunogenecity. Acta Biochim Biophys Sin. 2004;36:336–342. doi: 10.1093/abbs/36.5.336. [DOI] [PubMed] [Google Scholar]
- 289.Warmerdam P.A.M., Plaisance S., Vanderlick K., Vandervoort P., Brepoels K., Collen D. Elimination of a human T-cell region in staphylokinase by T-cell screening and computer modeling. Thromb Haemost. 2002;87:666–673. [PubMed] [Google Scholar]
- 290.Liu J., Wang Z., He J., Wang G., Zhang R., Zhao B. Effect of site-specific PEGylation on the fibrinolytic activity, immunogenicity, and pharmacokinetics of staphylokinase. Acta Biochim Biophys Sin. 2014;46:782–791. doi: 10.1093/abbs/gmu068. [DOI] [PubMed] [Google Scholar]
- 291.Liu R., Li D., Wang J., Qiu R., Lin Q., Zhang G. Preparation, characterization and in vitro bioactivity of N-terminally PEGylated staphylokinase dimers. Process Biochem. 2012;47:41–46. [Google Scholar]
- 292.Xu Y., Shi Y., Zhou J., Yang W., Bai L., Wang S. Structure-based antigenic epitope and PEGylation improve the efficacy of staphylokinase. Microb Cell Fact. 2017;16:197. doi: 10.1186/s12934-017-0801-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Xue X., Li D., Yu J., Ma G., Su Z., Hu T. Phenyl linker-induced dense peg conformation improves the efficacy of c-terminally monopegylated staphylokinase. Biomacromolecules. 2013;14:331–341. doi: 10.1021/bm301511w. [DOI] [PubMed] [Google Scholar]
- 294.Qi F., Hu C., Yu W., Hu T. Conjugation with eight-arm peg markedly improves the in vitro activity and prolongs the blood circulation of staphylokinase. Bioconjug Chem. 2018;29:451–458. doi: 10.1021/acs.bioconjchem.7b00770. [DOI] [PubMed] [Google Scholar]
- 295.Collen D., Sinnaeve P., Demarsin E., Moreau H., De Maeyer M., Jespers L. Polyethylene glycol–derivatized cysteine-substitution variants of recombinant staphylokinase for single-bolus treatment of acute myocardial infarction. Circulation. 2000;102:1766–1772. doi: 10.1161/01.cir.102.15.1766. [DOI] [PubMed] [Google Scholar]
- 296.Miele R.G., Prorok M., Costa V.A., Castellino F.J. Glycosylation of asparagine-28 of recombinant staphylokinase with high-mannose-type oligosaccharides results in a protein with highly attenuated plasminogen activator activity. J Biol Chem. 1999;274:7769–7776. doi: 10.1074/jbc.274.12.7769. [DOI] [PubMed] [Google Scholar]
- 297.Wang M., Wang Y., Wang J., Zou M., Liu S., Xu T. Construction and characterization of a novel staphylokinase variant with thrombin-inhibitory activity. Biotechnol Lett. 2009;31:1923–1927. doi: 10.1007/s10529-009-0094-2. [DOI] [PubMed] [Google Scholar]
- 298.Kowalski M., Brown G., Bieniasz M., Oszajca K., Chabielska E., Pietras T. Cloning and expression of a new recombinant thrombolytic and anthithrombotic agent - a staphylokinase variant. Acta Biochim Pol. 2009;56:41–53. [PubMed] [Google Scholar]
- 299.Chiou J.-F., Woon M.-D., Cheng S.-N., Hsu C.-H., Cherng S.-C., Hsieh F.-K. Staphylokinase-annexin XI chimera exhibited efficient in vitro thrombolytic activities. Biosci Biotechnol Biochem. 2007;71:1122–1129. doi: 10.1271/bbb.60279. [DOI] [PubMed] [Google Scholar]
- 300.Lian Q., Szarka S.J., Ng K.K.S., Wong S.L. Engineering of a staphylokinase-based fibrinolytic agent with antithrombotic activity and targeting capability toward thrombin-rich fibrin and plasma clots. J Biol Chem. 2003;278:26677–26686. doi: 10.1074/jbc.M303241200. [DOI] [PubMed] [Google Scholar]
- 301.Wu S.-C., Castellino F.J., Wong S.-L. A Fast-acting, modular-structured staphylokinase fusion with kringle-1 from human plasminogen as the fibrin-targeting domain offers improved clot lysis efficacy. J Biol Chem. 2003;278:18199–18206. doi: 10.1074/jbc.M210919200. [DOI] [PubMed] [Google Scholar]
- 302.Mannully S.T., Shanthi C., Pulicherla K.K. Lipid modification of staphylokinase and its implications on stability and activity. Int J Biol Macromol. 2019;121:1037–1045. doi: 10.1016/j.ijbiomac.2018.10.134. [DOI] [PubMed] [Google Scholar]
- 303.Cartwright T. The plasminogen activator of vampire bat saliva. Blood. 1974;43:317–326. [PubMed] [Google Scholar]
- 304.Hawkey C. Plasminogen activator in saliva of the vampire bat desmodus rotundus. Nature. 1966;211:434–435. doi: 10.1038/211434c0. [DOI] [PubMed] [Google Scholar]
- 305.Gardell S.J., Ramjit D.R., Stabilito I.I., Fujita T., Lynch J.J., Cuca G.C. Effective thrombolysis without marked plasminemia after bolus intravenous administration of vampire bat salivary plasminogen activator in rabbits. Circulation. 1991;84:244–253. doi: 10.1161/01.cir.84.1.244. [DOI] [PubMed] [Google Scholar]
- 306.Gardell S.J., Hare T.R., Bergum P.W., Cuca G.C., O’Neill-Palladino L., Zavodny S.M. Vampire bat salivary plasminogen activator is quiescent in human plasma in the absence of fibrin unlike human tissue plasminogen activator. Blood. 1990;76:2560–2564. [PubMed] [Google Scholar]
- 307.Mellott M.J., Stabilito I.I., Holahan M.A., Cuca G.C., Wang S., Li P. Vampire bat salivary plasminogen activator promotes rapid and sustained reperfusion without concomitant systemic plasminogen activation in a canine model of arterial thrombosis. Arterioscler Thromb J Vasc Biol. 1992;12:212–221. doi: 10.1161/01.atv.12.2.212. [DOI] [PubMed] [Google Scholar]
- 308.Witt W., Baldus B., Bringmann P., Cashion L., Donner P., Schleuning W.D. Thrombolytic properties of Desmodus rotundus (vampire bat) salivary plasminogen activator in experimental pulmonary embolism in rats. Blood. 1992;79:1213–1217. [PubMed] [Google Scholar]
- 309.Liberatore G.T., Samson A., Bladin C., Schleuning W.D., Medcalf R.L. Vampire bat salivary plasminogen activator (desmoteplase) - a unique fibrinolytic enzyme that does not promote neurodegeneration. Stroke. 2003;34:537–543. doi: 10.1161/01.str.0000049764.49162.76. [DOI] [PubMed] [Google Scholar]
- 310.Von Kummer R., Mori E., Truelsen T., Jensen J.S., Grønning B.A., Fiebach J.B. Desmoteplase 3 to 9 hours after major artery occlusion stroke. Stroke. 2016;47:2880–2887. doi: 10.1161/STROKEAHA.116.013715. [DOI] [PubMed] [Google Scholar]
- 311.Albers G.W., von Kummer R., Truelsen T., Jensen J.-K.S., Ravn G.M., Grønning B.A. Safety and efficacy of desmoteplase given 3–9 h after ischaemic stroke in patients with occlusion or high-grade stenosis in major cerebral arteries (DIAS-3): a double-blind, randomised, placebo-controlled phase 3 trial. Lancet Neurol. 2015;14:575–584. doi: 10.1016/S1474-4422(15)00047-2. [DOI] [PubMed] [Google Scholar]
- 312.Gardell S.J., Duong L.T., Diehl R.E., York J.D., Hare T.R., Register R.B. Isolation, characterization, and cDNA cloning of a vampire bat salivary plasminogen activator. J Biol Chem. 1989;264:17947–17952. [PubMed] [Google Scholar]
- 313.Krätzschmar J., Haendler B., Langer G., Boidol W., Bringmann P., Alagon A. The plasminogen activator family from the salivary gland of the vampire bat Desmodus rotundas: cloning and expression. Gene. 1991;105:229–237. doi: 10.1016/0378-1119(91)90155-5. [DOI] [PubMed] [Google Scholar]
- 314.Renatus M., Stubbs M.T., Huber R., Bode W., Bringmann P., Donner P. Catalytic domain structure of vampire bat plasminogen activator: a molecular paradigm for proteolysis without activation cleavage. Biochemistry. 1997;36:13483–13493. doi: 10.1021/bi971129x. [DOI] [PubMed] [Google Scholar]
- 315.Schleuning W.-D., Alagon A., Boidol W., Bringmann P., Petri T., Krätzschmar J. Plasminogen activators from the saliva of desmodus rotundus (common vampire bat): unique fibrin specificity. Ann N Y Acad Sci. 1992;667:395–403. doi: 10.1111/j.1749-6632.1992.tb51639.x. [DOI] [PubMed] [Google Scholar]
- 316.Gohlke M., Baude G., Nuck R., Grunow D., Kannicht C., Bringmann P. O-Linked L-Fucose Is present in desmodus rotundus salivary plasminogen activator. J Biol Chem. 1996;271:7381–7386. doi: 10.1074/jbc.271.13.7381. [DOI] [PubMed] [Google Scholar]
- 317.Gohlke M., Nuck R., Kannicht C., Grunow D., Baude G., Donner P. Analysis of site-specific N-glycosylation of recombinant desmodus rotundus salivary plasminogen activator rDSPAα1 expressed in Chinese hamster ovary cells. Glycobiology. 1997;7:67–77. doi: 10.1093/glycob/7.1.67. [DOI] [PubMed] [Google Scholar]
- 318.Bringmann P., Gruber D., Liese A., Toschi L., Kratzschmar J., Schleuning W. Structural features mediating fibrin selectivity of vampire bat plasminogen activators. J Biol Chem. 1995;270:25596–25603. doi: 10.1074/jbc.270.43.25596. [DOI] [PubMed] [Google Scholar]
- 319.Stewart R.J., Fredenburgh J.C., Weitz J.I. Characterization of the interactions of plasminogen and tissue and vampire bat plasminogen activators with fibrinogen, fibrin, and the complex of d-dimer noncovalently linked to fragment E. J Biol Chem. 1998;273:18292–18299. doi: 10.1074/jbc.273.29.18292. [DOI] [PubMed] [Google Scholar]
- 320.Toschi L., Bringmann P., Petri T., Donner P., Schleuning W.-D. Fibrin selectivity of the isolated protease domains of tissue-type and vampire bat salivary gland plasminogen activators. Eur J Biochem. 1998;252:108–112. doi: 10.1046/j.1432-1327.1998.2520108.x. [DOI] [PubMed] [Google Scholar]
- 321.Bergum P.W., Gardell S.J. Vampire bat salivary plasminogen activator exhibits a strict and fastidious requirement for polymeric fibrin as its cofactor, unlike human tissue-type plasminogen activator. a kinetic analysis. J Biol Chem. 1992;267:17726–17731. [PubMed] [Google Scholar]
- 322.Witt W., Kirchhoff D., Woy P., Zierz R., Bhargava A.S. Antibody formation and effects on endogenous fibrinolysis after repeated administration of dspaai in rats. Fibrinolysis. 1994;8:66. [Google Scholar]
- 323.Saadatirad A., Sardari S., Kazemali M., Zarei N., Davami F., Barkhordari F. Expression of a novel chimeric-truncated tpa in pichia pastoris with improved biochemical properties. Mol Biotechnol Totowa. 2014;56:1143–1150. doi: 10.1007/s12033-014-9794-5. [DOI] [PubMed] [Google Scholar]
- 324.Vanacek P., Sebestova E., Babkova P., Bidmanova S., Daniel L., Dvorak P. Exploration of enzyme diversity by integrating bioinformatics with expression analysis and biochemical characterization. ACS Catal. 2018;8:2402–2412. [Google Scholar]
- 325.Li Z., Chen X., Guo S., Zhang H., Dong H., Wu G. Engineering of Harobin for enhanced fibrinolytic activity obtained by random and site-directed mutagenesis. Protein Expr Purif. 2017;129:162–172. doi: 10.1016/j.pep.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 326.da CE Silva P.E., RC de Barros, WWC Albuquerque, RMP Brandão, Bezerra R.P., ALF Porto. In vitro thrombolytic activity of a purified fibrinolytic enzyme from Chlorella vulgaris. J Chromatogr B. 2018;1092:524–529. doi: 10.1016/j.jchromb.2018.04.040. [DOI] [PubMed] [Google Scholar]
- 327.Chen H., McGowan E.M., Ren N., Lal S., Nassif N., Shad-Kaneez F. Nattokinase: a promising alternative in prevention and treatment of cardiovascular diseases. Biomark Insights. 2018;13 doi: 10.1177/1177271918785130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Koyanagi K., Narasaki R., Yamamichi S., Suzuki E., Hasumi K. Mechanism of the action of SMTP-7, a novel small-molecule modulator of plasminogen activation. Blood Coagul Fibrinolysis. 2014;25:316–321. doi: 10.1097/MBC.0000000000000032. [DOI] [PubMed] [Google Scholar]
- 329.Hasegawa K., Koide H., Hu W., Nishimura N., Narasaki R., Kitano Y. Structure-activity relationships of 11 new congeners of the SMTP plasminogen modulator. J Antibiot (Tokyo) 2010;63:589–593. doi: 10.1038/ja.2010.101. [DOI] [PubMed] [Google Scholar]
- 330.Hasumi K., Yamamichi S., Harada T. Small-molecule modulators of zymogen activation in the fibrinolytic and coagulation systems. FEBS J. 2010;277:3675–3687. doi: 10.1111/j.1742-4658.2010.07783.x. [DOI] [PubMed] [Google Scholar]
- 331.Takayasu R., Hasumi K., Shinohara C., Endo A. Enhancement of fibrin binding and activation of plasminogen by staplabin through induction of a conformational change in plasminogen. FEBS Lett. 1997;418:58–62. doi: 10.1016/s0014-5793(97)01334-3. [DOI] [PubMed] [Google Scholar]
- 332.Suzuki E., Nishimura N., Yoshikawa T., Kunikiyo Y., Hasegawa K., Hasumi K. Efficacy of SMTP-7, a small-molecule anti-inflammatory thrombolytic, in embolic stroke in monkeys. Pharmacol Res Perspect. 2018;6(UNSP) doi: 10.1002/prp2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Shibata K., Hashimoto T., Hasumi K., Honda K., Nobe K. Evaluation of the effects of a new series of SMTPs in the acetic acid-induced embolic cerebral infarct mouse model. Eur J Pharmacol. 2018;818:221–227. doi: 10.1016/j.ejphar.2017.10.055. [DOI] [PubMed] [Google Scholar]
- 334.Korin N., Kanapathipillai M., Matthews B.D., Crescente M., Brill A., Mammoto T. Shear-activated nanotherapeutics for drug targeting to obstructed blood vessels. Science. 2012;337:738–742. doi: 10.1126/science.1217815. [DOI] [PubMed] [Google Scholar]
- 335.Liu S., Feng X., Jin R., Li G. Tissue plasminogen activator-based nanothrombolysis for ischemic stroke. Expert Opin Drug Deliv. 2018;15:173–184. doi: 10.1080/17425247.2018.1384464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Koudelka S., Mikulik R., Mašek J., Raška M., Turánek Knotigová P., Miller A.D. Liposomal nanocarriers for plasminogen activators. J Control Release. 2016;227:45–57. doi: 10.1016/j.jconrel.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 337.Chapurina Y.E., Drozdov A.S., Popov I., Vinogradov V.V., Dudanov I.P., Vinogradov V.V. Streptokinase@alumina nanoparticles as a promising thrombolytic colloid with prolonged action. J Mater Chem B. 2016;4:5921–5928. doi: 10.1039/c6tb01349j. [DOI] [PubMed] [Google Scholar]
- 338.Pitek A.S., Wang Y., Gulati S., Gao H., Stewart P.L., Simon D.I. Elongated plant virus-based nanoparticles for enhanced delivery of thrombolytic therapies. Mol Pharm. 2017;14:3815–3823. doi: 10.1021/acs.molpharmaceut.7b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Fernandes L.F., Bruch G.E., Massensini A.R., Frezard F. Recent advances in the therapeutic and diagnostic use of liposomes and carbon nanomaterials in ischemic stroke. Front Neurosci. 2018;12:453. doi: 10.3389/fnins.2018.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Mukherjee P., Leman L.J., Griffin J.H., Ghadiri M.R. Design of a DNA-programmed plasminogen activator. J Am Chem Soc. 2018;140:15516–15524. doi: 10.1021/jacs.8b10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Bjerregaard N., Bøtkjær K.A., Helsen N., Andreasen P.A., Dupont D.M. Tissue-type plasminogen activator-binding RNA aptamers inhibiting low-density lipoprotein receptor family-mediated internalisation. Thromb Haemost. 2015;114:139–149. doi: 10.1160/TH14-08-0686. [DOI] [PubMed] [Google Scholar]
- 342.Zeitelhofer M., Li H., Adzemovic M.Z., Nilsson I., Muhl L., Scott A.M. Preclinical toxicological assessment of a novel monoclonal antibody targeting human platelet-derived growth factor CC (PDGF-CC) in PDGF-CChum mice. Plos One. 2018;13 doi: 10.1371/journal.pone.0200649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Kikuchi K., Setoyama K., Tanaka E., Otsuka S., Terashi T., Nakanishi K. Uric acid enhances alteplase-mediated thrombolysis as an antioxidant. Sci Rep. 2018;8:15844. doi: 10.1038/s41598-018-34220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.dela Pena I.C., Yang S., Shen G., Liang H.F., Solak S., Borlongan C.V. Extension of tissue plasminogen activator treatment window by granulocyte-colony stimulating factor in a thromboembolic rat model of stroke. Int J Mol Sci. 2018;19:1635. doi: 10.3390/ijms19061635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Niego B., Lee N., Larsson P., de Silva T.M., Au A.E.-L., McCutcheon F. Selective inhibition of brain endothelial Rhokinase-2 provides optimal protection of an in vitro blood-brain barrier from tissue-type plasminogen activator and plasmin. Plos One. 2017;12 doi: 10.1371/journal.pone.0177332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Niego B., Broughton B.R.S., Ho H., Sobey C.G., Medcalf R.L. LDL receptor blockade reduces mortality in a mouse model of ischaemic stroke without improving tissue-type plasminogen activator-induced brain haemorrhage: towards pre-clinical simulation of symptomatic ICH. Fluids Barriers Cns. 2017;14:33. doi: 10.1186/s12987-017-0081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Knecht T., Story J., Liu J., Davis W., Borlongan C.V., dela Pena I.C. Adjunctive therapy approaches for ischemic stroke: innovations to expand time window of treatment. Int J Mol Sci. 2017;18:2756. doi: 10.3390/ijms18122756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Kanazawa M., Takahashi T., Nishizawa M., Shimohata T. Therapeutic strategies to attenuate hemorrhagic transformation after tissue plasminogen activator treatment for acute ischemic stroke. J Atheroscler Thromb. 2017;24:240–253. doi: 10.5551/jat.RV16006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Shimamura M., Nakagami H., Shimizu H., Mukai H., Watanabe R., Okuzono T. Development of a novel RANKL-based peptide, microglial healing peptide1-AcN (MHP1-AcN), for treatment of ischemic stroke. Sci Rep. 2018;8:17770. doi: 10.1038/s41598-018-35898-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Vercauteren E., Gils A., Declerck P. Thrombin activatable fibrinolysis inhibitor: a putative target to enhance fibrinolysis. Semin Thromb Hemost. 2013;39:365–372. doi: 10.1055/s-0033-1334488. [DOI] [PubMed] [Google Scholar]
- 351.Schreuder H., Liesum A., Loenze P., Stump H., Hoffmann H., Schiell M. Isolation, co-crystallization and structure-based characterization of anabaenopeptins as highly potent inhibitors of activated thrombin activatable fibrinolysis inhibitor (TAFIa) Sci Rep. 2016;6:32958. doi: 10.1038/srep32958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Halland N., Broenstrup M., Czech J., Czechtizky W., Evers A., Follmann M. novel small molecule inhibitors of activated thrombin activatable fibrinolysis inhibitor (tafia) from natural product anabaenopeptin. J Med Chem. 2015;58:4839–4844. doi: 10.1021/jm501840b. [DOI] [PubMed] [Google Scholar]
- 353.Wyseure T., Rubio M., Denorme F., de Lizarrondo S.M., Peeters M., Gils A. Innovative thrombolytic strategy using a heterodimer diabody against TAFI and PAI-1 in mouse models of thrombosis and stroke. Blood. 2015;125:1325–1332. doi: 10.1182/blood-2014-07-588319. [DOI] [PubMed] [Google Scholar]
- 354.Denorme F., Wyseure T., Peeters M., Vandeputte N., Gils A., Deckmyn H. Inhibition of thrombin-activatable fibrinolysis inhibitor and plasminogen activator inhibitor-1 reduces ischemic brain damage in mice. Stroke. 2016;47:2419–2422. doi: 10.1161/STROKEAHA.116.014091. [DOI] [PubMed] [Google Scholar]
- 355.Singh S., Houng A., Reed G.L. Releasing the brakes on the fibrinolytic system in pulmonary emboli unique effects of plasminogen activation and alpha 2-antiplasmin inactivation. Circulation. 2017;135:1011–1020. doi: 10.1161/CIRCULATIONAHA.116.024421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Urano T., Suzuki Y. Thrombolytic therapy targeting alpha 2-antiplasmin. Circulation. 2017;135:1021–1023. doi: 10.1161/CIRCULATIONAHA.117.026884. [DOI] [PubMed] [Google Scholar]
- 357.Baluta M.M., Vintila M.M. PAI-1 inhibition – another therapeutic option for cardiovascular protection. Mædica. 2015;10:147–152. [PMC free article] [PubMed] [Google Scholar]
- 358.Maheshwari N., Kantipudi S., Maheshwari A., Arora K., Kwatra V.N., Sahni G. Amino-terminal fusion of epidermal growth factor 4,5,6 domains of human thrombomodulin on streptokinase confers anti-reocclusion characteristics along with plasmin-mediated clot specificity. Plos One. 2016;11 doi: 10.1371/journal.pone.0150315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Absar S., Gupta N., Nahar K., Ahsan F. Engineering of plasminogen activators for targeting to thrombus and heightening thrombolytic efficacy. J Thromb Haemost. 2015;13:1545–1556. doi: 10.1111/jth.13033. [DOI] [PubMed] [Google Scholar]
- 360.Armstead W.M., Hekierski H., Yarovoi S., Higazi A.A.-R., Cines D.B. tPA variant tPA-A(296-299) Prevents impairment of cerebral autoregulation and necrosis of hippocampal neurons after stroke by inhibiting upregulation of ET-1. J Neurosci Res. 2018;96:128–137. doi: 10.1002/jnr.24112. [DOI] [PubMed] [Google Scholar]
- 361.Shafiee F., Moazen F., Rabbani M., Mohammad Sadeghi H.M. Expression and activity evaluation of reteplase in Escherichia coli TOP10. J Paramed Sci. 2015;6:58–64. [Google Scholar]
- 362.Zhuo X.-F., Zhang Y.-Y., Guan Y.-X., Yao S.-J. Co-expression of disulfide oxidoreductases DsbA/DsbC markedly enhanced soluble and functional expression of reteplase in Escherichia coli. J Biotechnol. 2014;192:197–203. doi: 10.1016/j.jbiotec.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 363.Majidzadeh-A K., Khalaj V., Fatemeh D., Mahdi H., Farzaneh B., Ahmad A. Cloning and expression of functional full-length human tissue plasminogen activator in pichia pastoris. Appl Biochem Biotechnol. 2010;162:2037–2048. doi: 10.1007/s12010-010-8979-z. [DOI] [PubMed] [Google Scholar]
- 364.Wei Z., Wang Y., Li G., Li X., Liu D. Optimized gene synthesis, expression and purification of active salivary plasminogen activator α2 (DSPAα2) of desmodus rotundus in Pichia pastoris. Protein Expr Purif. 2008;57:27–33. doi: 10.1016/j.pep.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 365.Faraji H., Ramezani M., Sadeghnia H.R., Abnous K., Soltani F., Mashkani B. High-level expression of a biologically active staphylokinase in Pichia pastoris. Prep Biochem Biotechnol. 2017;47:379–387. doi: 10.1080/10826068.2016.1252924. [DOI] [PubMed] [Google Scholar]
- 366.Fathi-Roudsari M., Maghsoudi N., Maghsoudi A., Niazi S., Soleiman M. Auto-induction for high level production of biologically active reteplase in Escherichia coli. Protein Expr Purif. 2018;151:18–22. doi: 10.1016/j.pep.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 367.Wang H., Wang F., Wei D. Impact of oxygen supply on rtPA expression in Escherichia coli BL21 (DE3): ammonia effects. Appl Microbiol Biotechnol Heidelb. 2009;82:249–259. doi: 10.1007/s00253-008-1756-z. [DOI] [PubMed] [Google Scholar]
- 368.Mousavi S.B., Fazeli A., Shojaosadati S.A., Fazeli M.R., Hashemi-Najafabadi S. Purification and efficient refolding process for recombinant tissue-type plasminogen activator derivative (reteplase) using glycerol and Tranexamic acid. Process Biochem. 2017;53:135–144. [Google Scholar]
- 369.Mousavi S.B., Fazeli A., Shojaosadati S.A., Fazeli M.R. Development of a two-step refolding method for reteplase, a rich disulfide-bonded protein. Process Biochem. 2018;74:94–102. [Google Scholar]
- 370.Liu H., Zhou X., Zhang Y. A comparative investigation on different refolding strategies of recombinant human tissue-type plasminogen activator derivative. Biotechnol Lett. 2006;28:457–463. doi: 10.1007/s10529-006-0001-z. [DOI] [PubMed] [Google Scholar]
- 371.Rahimpour A., Ahani R., Najaei A., Adeli A., Barkhordari F., Mahboudi F. Development of genetically modified chinese hamster ovary host cells for the enhancement of recombinant tissue plasminogen activator expression. Malays J Med Sci Kelant. 2016;23:6–13. [PMC free article] [PubMed] [Google Scholar]
- 372.Hahn B.-S., Sim J.-S., Kim H.-M., Ahn M.-Y., Pak H.-K., Kim N.-A. Expression and characterization of human tissue-plasminogen activator in transgenic tobacco plants. Plant Mol Biol Report. 2008;27:209. [Google Scholar]
- 373.Nabiabad H.S., Piri K., Amini M. Expression of active chimeric-tissue plasminogen activator in tobacco hairy roots, identification of a DNA aptamer and purification by aptamer functionalized-MWCNTs chromatography. Protein Expr Purif. 2018;152:137–145. doi: 10.1016/j.pep.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 374.Song S., Ge X., Cheng Y., Lu R., Zhang T., Yu B. High-level expression of a novel recombinant human plasminogen activator (rhPA) in the milk of transgenic rabbits and its thrombolytic bioactivity in vitro. Mol Biol Rep. 2016;43:775–783. doi: 10.1007/s11033-016-4020-0. [DOI] [PubMed] [Google Scholar]
- 375.Kim S.-R., Sim J.-S., Ajjappala H., Kim Y.-H., Hahn B.-S. Expression and large-scale production of the biochemically active human tissue-plasminogen activator in hairy roots of Oriental melon (Cucumis melo) J Biosci Bioeng. 2012;113:106–111. doi: 10.1016/j.jbiosc.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 376.He Z., Lu R., Zhang T., Jiang L., Zhou M., Wu D. A novel recombinant human plasminogen activator: Efficient expression and hereditary stability in transgenic goats and in vitro thrombolytic bioactivity in the milk of transgenic goats. Plos One. 2018;13 doi: 10.1371/journal.pone.0201788. [DOI] [PMC free article] [PubMed] [Google Scholar]