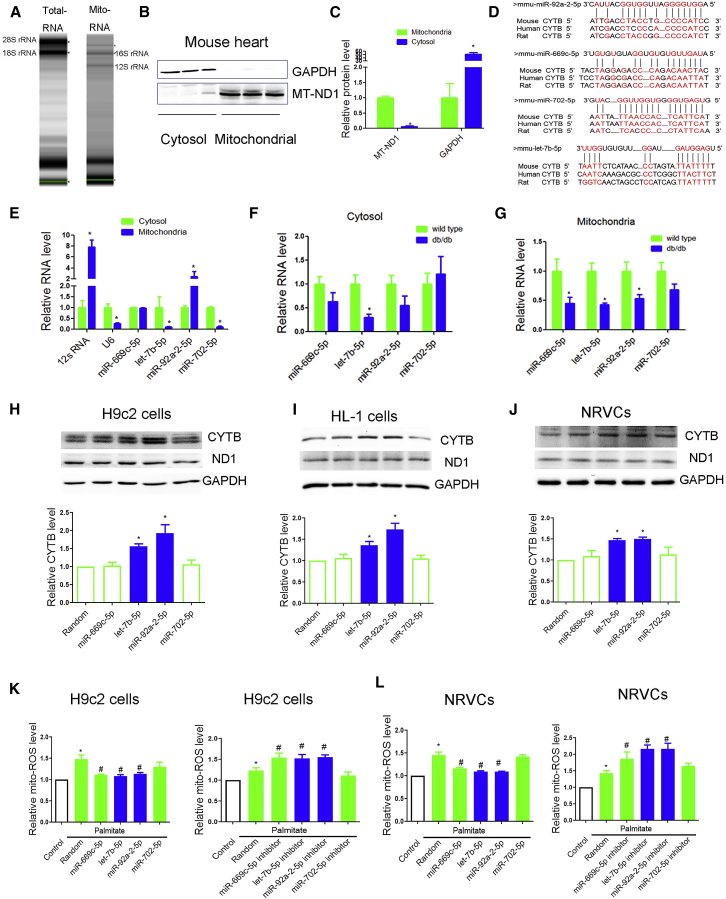
Figure 3.
miRNA Profile Was Performed in Cytoplasmic and Mitochondrial Components of db/db Mouse Heart
(A) Gel electrophoresis was used to analyze the peak of mitochondrial RNA versus total heart. (B and C) Mitochondrial ND1 and GAPDH in mitochondria and cytosol analyzed by western blot (B) were evaluated for significance using Student’s t test (C). (D) Sequence alignment of miR-92a-2-5p, let-7b-5p, miR-669c-5p, and miR-702 on the Cytb mRNA from different organisms. (E) Relative expression of selected miRNAs in mitochondria in comparison with cytosol were detected by quantitative real-time PCR. 100 ng of RNA from cytosol and mitochondria were reverse transcribed and used as template for quantitative real-time PCR. The expression of RNA in mitochondria was represented using a fold change relative to the value 1 of the cytosol. n = 3; *p < 0.05 versus cytosol. (F and G) Relative expression of selected miRNAs in cytosol (F) and mitochondria (G) of db/db mouse heart was detected by quantitative real-time PCR. Cytoplasmic RNAs were normalized to GAPDH mRNA and mitochondrial RNAs to 12 s rRNA. n = 8; *p < 0.05 versus wild-type. (H–J) Effects of miR-92a-2-5p, let-7b-5p, miR-669c-5p, and miR-702 on mitochondrial subunits at the protein levels in H9c2 cardiomyocytes (H), HL-1 cardiomyocytes (I), and NRVCs (J) were detected by western blot. n = 3; *p < 0.05 versus random. (K and L) Effects of miR-92a-2-5p, let-7b-5p, miR-669c-5p, and miR-702 on mitochondrial ROS under palmitate treatment were detected by MitoSOX red probe in H9c2 cardiomyocytes (K) and NRVCs (L). Cells were incubated with palmitate (0.25 mmol/L) for 24 h; n = 3; results from controls were set to 1; *p < 0.05 versus control, #p < 0.05 versus palmitate-random.