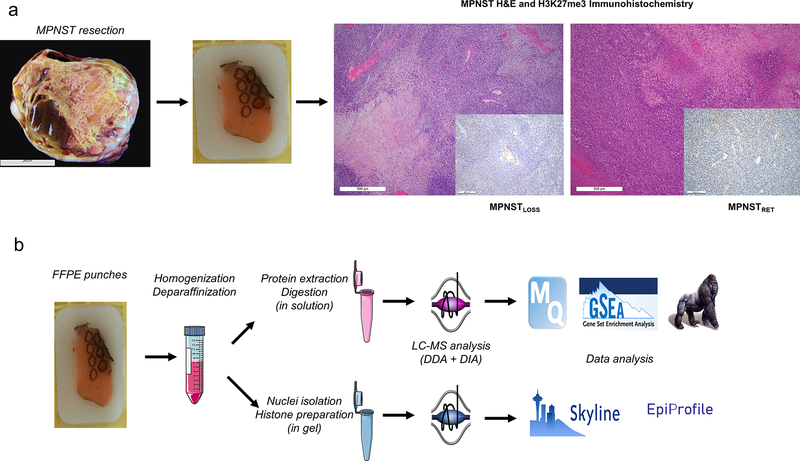
Figure 1.
Workflow for parallel proteomic and histone analysis of MPNSTLOSS and MPNSTRET. A, MPNST are resected (gross image of representative tumor at right) and processed according to standard pathology procedures. Representative sections from tumor blocks from pathology archives were reviewed on hematoxylin and eosin (H&E) histologic sections to identify the regions of interest. IHC (inset) on adjacent sections allowed for separation of MPNST tumors into MPNSTLOSS (left) and MPNSTRET (right) on the basis of H3K27me3 staining. MPNSTLOSS were defined by global loss of H3K27me3 nuclear staining, with retained internal control staining in nontumor tissue (blood vessel, center of IHC inset). MPNSTRET were defined by any degree of retained staining in tumor nuclei. B, From blocks identified in A, tissue was isolated from representative cores of FFPE tissues, and total protein was extracted. A portion of the total protein was processed in solution for MS and proteomic analysis using standard processing and analysis software (top), and a portion was sampled for SDS-PAGE, from which the histone bands were excised for in-gel digestion, derivatization, and histone PTM analysis using distinct analysis software (bottom).