Figure 5.
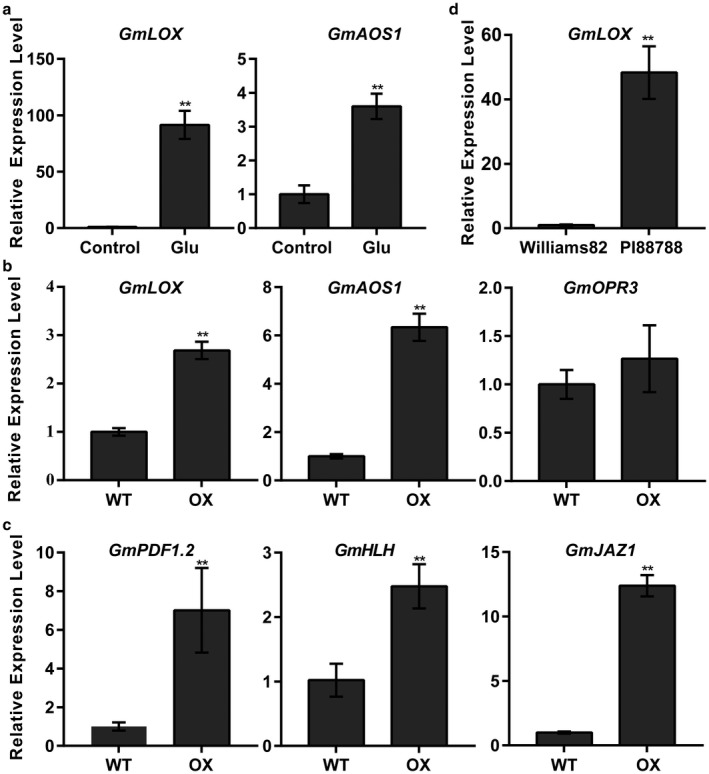
Expression of a set of jasmonic acid (JA) biosynthesis and JA‐responsive genes. (a) JA biosynthesis genes expressed in the roots of glutamic acid (Glu) ‐treated soybean. Roots were treated with quarter‐strength Murashige and Skoog medium that either lacked nitrogen (control) or was supplemented with 5 mm Glu for 24 h. JA biosynthesis genes (b) and JA‐responsive genes (c) expressed in the roots of the wild‐type (cultivar Tianlong 1) and Rhg1‐GmAAT‐overexpressing (Rhg1‐GmAAT‐OX) line gm‐3. (d) JA biosynthesis genes expressed in the roots of Williams 82 and PI88788. The expression levels of the selected genes were assayed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and were normalized to SKIP16. The values are the means ± standard deviations (SDs) (n = 3). **0.01 < P < 0.05, **P < 0.01 (multiple t‐test followed by the Holm–Sidak post hoc test). WT, wild‐type; OX, Rhg1‐GmAAT‐OX line gm‐3.
