Summary
Both Solanum tuberosum and Ralstonia solanacearum phylotype IIB originated in South America and share a long‐term co‐evolutionary history. However, our knowledge of potato bacterial wilt pathogenesis is scarce as a result of the technical difficulties of potato plant manipulation. Thus, we established a multiple screening system (virulence screen of effector mutants in potato, growth inhibition of yeast and transient expression in Nicotiana benthamiana) of core type III effectors (T3Es) of a major potato pathovar of phylotype IIB, to provide more research perspectives and biological tools. Using this system, we identified four effectors contributing to virulence during potato infection, with two exhibiting multiple phenotypes in two other systems, including RipAB. Further study showed that RipAB is an unknown protein with a nuclear localization signal (NLS). Furthermore, we generated a ripAB complementation strain and transgenic ripAB‐expressing potato plants, and subsequent virulence assays confirmed that R. solanacearum requires RipAB for full virulence. Compared with wild‐type potato, transcriptomic analysis of transgenic ripAB‐expressing potato plants showed a significant down‐regulation of Ca2+ signalling‐related genes in the enriched Plant–Pathogen Interaction (PPI) gene ontology (GO) term. We further verified that, during infection, RipAB is required for the down‐regulation of four Ca2+ sensors, Stcml5, Stcml23, Stcml‐cast and Stcdpk2, and a Ca2+ transporter, Stcngc1. Further evidence showed that the immune‐associated reactive oxygen species (ROS) burst is attenuated in ripAB transgenic potato plants. In conclusion, a systematic screen of conserved R. solanacearum effectors revealed an important role for RipAB, which interferes with Ca2+‐dependent gene expression to promote disease development in potato.
Keywords: type III effector, Ralstonia solanacearum, plant immunity, potato, calcium signalling
Introduction
Potato (Solanum tuberosum L.) is the third most important food crop worldwide. More than a billion people consume potato, and its production reached 382 million metric tons in 2014 (Kadota et al., 2016; McIntyre, 2003). However, potato production and quality are severely constrained by numerous diseases, such as bacterial wilt, caused by Ralstonia solanacearum (Champoiseau et al., 2009; Wullings et al., 1998). The major pathovar infecting potato belongs to phylotype IIB, which is widely distributed worldwide (Fegan et al., 2005; Hayward, 2003; Janse, 1996; Seal et al., 1999). Phylotype IIB pathogens have adapted to cool temperatures and have migrated worldwide, probably through contaminated potato tubers (Hayward, 1964; Hooker, 1981; Huerta et al., 2015; Wang et al., 2017; Wicker et al., 2012). Even though potato and R. solanacearum have a long‐term co‐evolutionary history, very little is known about the interaction between them, as a result of both a lack of resistance materials and technical barriers involving potato genetic manipulation (Chen et al., 2013, 2016). New strategies and biological tools are seriously needed for research and disease control.
Type III effectors (T3Es) are key virulence factors of R. solanacearum to counteract host immunity (Deslandes and Genin, 2014; Macho and Zipfel, 2015; Peeters et al., 2013b), and T3Es from different bacterial pathogens have also been shown to manipulate other cellular functions independent of immunity (Macho, 2016). Therefore, the use of effectors as molecular probes to detect potato immunity would offer a new perspective to study host resistance in potato. To date, this strategy has been broadly used in many studies of plant pathogenesis (Block and Alfano, 2011; Greenberg and Vinatzer, 2003; Kay and Bonas, 2009; Lindeberg et al., 2012; Mudgett, 2005; Zhou and Chai, 2008). With the development of genomics, an increasing number of R. solanacearum strains have been sequenced, and a plethora of effectors have been identified (Burstein et al., 2009, Peeters et al., 2013a; Petnickiocwieja et al., 2002). Currently, more than 100 effectors have been reported in the R. solanacearum species complex (Deslandes and Genin, 2014; Lonjon et al., 2018; Peeters et al., 2013a). Hence, a larger scale functional screening of effectors is essential for both the provision of more basic information on the effector inventory and acceleration of research on host resistance.
Ralstonia solanacearum can infect more than 200 different species of plant. Each individual R. solanacearum strain harbours a large effector inventory (60–70) that can respond to various host environments (Genin and Denny, 2012). Ralstonia solanacearum may require different combinations of effectors to infect specific hosts. Under these circumstances, the virulence screening of effector mutants is necessary for the study of specific host interactions and to prioritize effector studies amongst a large inventory.
Infection assays using effector mutants provide a direct proof of their contribution to virulence, but the detection of reduced virulence of individual effector mutants is difficult because of functional redundancy (Genin and Denny, 2012; Macho et al., 2010; Poueymiro et al., 2014). Owing to the conserved biochemical function of Gram‐negative bacterial effectors in eukaryotic environments, heterologous expression in yeast offers an effective and promising tool for whole‐genome functional screening and biochemical studies (Bosis et al., 2011; Curak et al., 2009). The function of numerous bacterial effectors has been revealed using yeast as a model organism (Fujiwara et al., 2016; Mukaihara et al., 2016; Salomon et al., 2011; Turgeon et al., 2009). Transient expression in Nicotiana benthamiana is also an ideal system for genetic investigations of plant interactions with pathogens, such as oomycetes, fungi and bacteria (Bombarely et al., 2012; Goodin et al., 2008; Oh et al., 2009; Poueymiro et al., 2009). All of these methods combined would provide versatile tools and perspectives to benefit our research.
Compared with Pseudomonas and Xanthomonas, Ralstonia harbours a larger effector repertoire of approximately 110 members (Lonjon et al., 2018; Peeters et al., 2013a). To date, the biochemical functions of a small number of these effectors and their targets have been described. RipP2 (PopP2) functions as an acetyltransferase that targets WRKY transcription factors (Le Roux et al., 2015; Sarris et al., 2015). RipAY acts as a γ‐glutamyl cyclotransferase, altering intracellular redox homeostasis in planta (Fujiwara et al., 2016; Mukaihara et al., 2016; Sang et al., 2018). RipG family members can interfere with E3 ligase complexes (Angot et al., 2006; Remigi et al., 2011; Wang et al., 2015). RipTPS acts as a trehalose‐6‐phosphate (T6P) synthase, promoting the biosynthesis of T6P, and thus altering the crosstalk between plant metabolism and development (Poueymiro et al., 2014). RipAK can suppress the plant hypersensitive response (HR) by the inhibition of catalases (Sun et al., 2017). However, the function of most Ralstonia effectors is not yet understood.
In this work, we used a multiple screening system, involving the virulence screening of effector mutants in potato, yeast growth inhibition and transient expression in tobacco, to identify R. solanacearum virulence effectors. Using this screening system, we identified a highly relevant T3E, RipAB, which is the first reported effector that can suppress the Ca2+ signalling pathway at the mRNA level to promote the infection of R. solanacearum in potato plants.
Results
Multiple functional screening reveals T3Es required for R. solanacearum virulence
To establish the multiple screening system, we first analysed the effector repertoire of 13 sequenced phylotype IIB strains (UW551, 23_10BR, CFBP1416, CFBP6783, CIP417, IBSBF1503, NCPPB909, NCPPB_282, P673, Po82, UW163, UW179 and UY031) in the R. solanacearum T3E database (Peeters et al., 2013a). Thirty‐three effectors, which were present more than 10 times in the 13 sequenced strains, were selected as core effectors. Previous studies have shown that 20 and four core effectors are up‐regulated inside plant xylem and root tissues, respectively, in comparison with their expression in rich medium (Jacobs et al., 2013, Puigvert et al., 2017). This indicates that most of these core effectors are associated with plant infection processes (Table 1). To reveal the virulence contribution of these core effectors, three different functional screenings were performed, including virulence assays of mutants on potato, growth inhibition of yeast and transient expression in N. benthamiana.
Table 1.
The virulence functional screening of Ralstonia solanacearum effectors.
| Name† | Description‡ | Presence in phylotype IIB (%)§ | Expression level¶ | Yeast growth inhibition†† | Cell death in Nicotiana‡‡ | Disease index in potato§§ | |||
|---|---|---|---|---|---|---|---|---|---|
|
Stem vs. Medium (log2 FC) |
Root vs. Medium (log2 FC) |
30 °C | 0.5M NaCl | N. benthamiana | Disease index | Significance | |||
| ripA2 | AWR2 | 100 | 2.92 | ns | None | None | None | – | – |
| ripA5_2 | AWR5 | 77 | ns | ns | Strong | Strong | – | 0.9219 + 0.1197 | ns |
| ripAB | NLS‐harbouring protein | 100 | 27.80 | 4.27 | Weak | Weak | Strong | 0.7813 + 0.2016 | ** |
| ripAC | LRR domain | 92 | 17.11 | 3.35 | None | None | – | 0.9063 + 0.125 | ns |
| ripAD | – | 92 | 3.87 | ns | None | None | None | 0.8281 + 0.1505 | ns |
| ripAE | Putative acetyltransferase | 77 | 10.09 | ns | None | None | None | 0.9063 + 0.125 | ns |
| ripAI | – | 92 | 3.51 | ns | None | None | None | – | – |
| ripAJ | – | 92 | ns | ns | None | None | None | 0.9531 + 0.1008 | ns |
| ripAM | – | 85 | 2.07 | ns | None | None | None | 0.75 + 0.2887 | *** |
| ripAN | – | 85 | 3.58 | ns | None | Strong | – | 0.7188 + 0.2213 | **** |
| ripAP | – | 77 | ns | ns | None | None | None | 0.9531 + 0.1008 | ns |
| ripAR | Ubiquitin ligase domain | 85 | ns | ns | None | None | None | – | – |
| ripAT | – | 77 | 3.09 | ns | None | None | None | – | – |
| ripAV | – | 92 | ns | ns | None | Strong | None | – | – |
| ripAY | Glutamyl cyclotransferase | 100 | ns | ns | Strong | Strong | None | – | – |
| ripB | Nucleoside ribohydrolase | 85 | ns | ns | None | None | – | – | – |
| ripBH | – | 77 | 3.03 | ns | Strong | Strong | Strong | 0.7656 + 0.17 | ** |
| ripC1 | HAD‐like phosphatase | 85 | 9.87 | ns | None | None | None | – | – |
| ripD | – | 77 | 23.66 | 3.76 | None | Weak | None | – | – |
| ripE1 | – | 77 | ns | ns | None | None | – | – | – |
| ripE2 | – | 85 | 6.40 | ns | None | None | None | – | – |
| ripF1 | T3SS translocator | 77 | 23.73 | 3.94 | None | None | None | – | – |
| ripF2 | T3SS translocator | 92 | 2.25 | ns | None | None | Weak | 0.9531 + 0.1008 | ns |
| ripG2 | F‐box LRR GALA2 | 77 | 2.30 | ns | None | None | None | 0.9844 + 0.0625 | ns |
| ripG6 | F‐box LRR GALA6 | 85 | ns | ns | None | None | None | 0.8906 + 0.1281 | ns |
| ripG7 | F‐box LRR GALA7 | 92 | 17.54 | ns | None | None | None | 0.8906 + 0.1281 | ns |
| ripH1 | – | 77 | 2.09 | ns | None | None | None | 0.9219 + 0.1197 | ns |
| ripN | Nudix hydrolase | 77 | ns | ns | None | None | – | – | – |
| ripO1 | – | 92 | ns | ns | None | None | None | 0.8906 + 0.1281 | ns |
| ripR | – | 100 | ns | ns | None | None | None | 0.9219 + 0.1197 | ns |
| ripTPS | Trehalose phosphate synthase | 92 | ns | −2.61 | None | None | None | 0.9688 + 0.08539 | ns |
| ripU | – | 100 | 3.13 | ns | None | None | None | – | – |
| ripV1 | Ubiquitin ligase domain | 100 | 2.97 | ns | Weak | Strong | Weak | – | – |
| UW551 | – | – | – | – | Weak | 0.9531 + 0.1008 | |||
HAD, haloacid dehydrogenase; LRR, leucine‐rich repeat; NLS, nuclear localization signal; ns, not significant; T3SS, type III secretion system.
†‡Effector names and functional descriptions are from the type III effector (T3E) database (Peeters et al., 2013a).
§The effector presence percentage was calculated from 13 strains in phylotype IIB.
¶The transcriptomic data are from Jacobs et al. (2013) and Puigvert et al. (2017), and the values are log2 FC (fold change).
††Yeast growth inhibition screenings were performed under two conditions (normal conditions and salt stress conditions with 0.5 m NaCl). The images were obtained 2 days after incubation. Two standards (weak/strong) were used to evaluate yeast, as depicted in Fig. S5.
‡‡Effectors developing cell death phenotypes in Nicotiana benthamiana. Cell death triggered by effectors in N. benthamiana was evaluated at 96 h post‐inoculation (hpi); two standards (weak/strong) were used to evaluate cell death, as depicted in Fig. S6.
§§Virulence screening of R. solanacearum UW551 effector mutants in potato. The disease indices were recorded at 14 days post‐inoculation (dpi) [means + standard deviations (SDs), n = 4, grey shading indicates P < 0.01, one‐way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test). The table cells with grey shading indicate significant differences from the control.
To investigate which effectors are important for R. solanacearum potato infection, we generated a collection of effector knockout mutant. Twenty‐four core effectors were successfully mutated in the reference UW551 strain via allelic exchange with spectinomycin resistance (Smr) cassettes (Jacobs et al., 2013). Compared with the wild‐type (WT) strain, six mutants were discarded because of distinguishable differences in motility assays and growth assays in rich medium (Figs S1 and S2, see Supporting Information). The remaining 18 mutants were subjected to virulence assays using a susceptible potato accession, C9701, as described previously (Chen et al., 2013). Compared with the UW551 WT control, four of these effector mutants (ripAM, ripAB, ripAN and ripBH) showed a significant reduction in virulence (Table 1; Figs S3 and S4, see Supporting Information). These data indicate that each of these four effectors is required for infection of the susceptible potato accession C9701.
Furthermore, we used a yeast heterologous expression system to investigate the virulence function of the core effectors. We cloned 33 core effectors into a galactose‐inducible vector and transformed them into the yeast strain BY4741. The transformants were grown on both suppression and induction medium under two different conditions (with and without 0.5 m NaCl). The results showed that four effectors, RipA5, RipBH, RipV1 and RipAY, strongly inhibited yeast growth under the two conditions. However, RipAB, RipAN, RipAV and RipD caused a relatively weak growth inhibition (Table 1; Fig. S5). Together, these data indicate that these effectors interfere with basic cellular functions in yeast.
Interestingly, using transient expression in N. benthamiana, we observed that four effectors (RipAB, RipV1, RipBH and RipF2) triggered cell death at 96 h post‐inoculation (hpi) (Table 1; Fig. S6, see Supporting Information). Compared with RipV1 and RipF2, RipBH and RipAB triggered a stronger tissue necrosis (Fig. S7, see Supporting Information). As a control for the immune elicitor that induces cell death, we used the Phytophthora infestans elicitor INF1 (Du et al., 2015). However, unlike cell death triggered by INF1, the cell death triggered by these four effectors seemed to be independent of the SGT1‐, EDS1‐, NDR1‐, HSP70‐ and MEKK2‐mediated immune signalling pathways, as virus‐induced gene silencing (VIGS) of these genes did not affect the cell death triggered by these effectors (Figs S8 and S9, see Supporting Information). These findings suggest that the cell death triggered by these effectors may be independent of the signalling pathways mediated by these genes.
As RipAB is required for full virulence during potato infection, induces cell death in N. benthamiana and inhibits yeast growth, we selected RipAB as our primary research object.
RipAB is a type III‐secreted effector in vivo
Previous studies have shown that RipAB is a type III‐secreted effector in vitro (Gueneron et al., 2000; Lonjon et al., 2015). To test the secretion of RipAB during potato infection, an adenylate cyclase (cyaA) assay was performed as described previously (Poueymiro et al., 2014), with some modifications. We generated an R. solanacearum expression vector (pABsscyaA) using a fragment of 100 amino acids of the RipAB N‐terminal type III secretion signal fused to cyaA protein (Fig. 1a). We then transformed the expression vector and an empty vector (pABcyaA) into both UW551 and the type III secretion system (T3SS) mutant ΔHrcV. If it is translocated by the T3SS, cyaA would significantly increases the concentration of cyclic adenosine monophosphate (cAMP) in the cytoplasm. The concentration of cAMP was monitored at 7 days post‐inoculation (dpi). We observed that the concentration of cAMP in plants inoculated with UW551 (pABsscyaA) was significantly higher than that in plants inoculated with ΔHrcV (pABsscyaA), UW551(pABcyaA), ΔhrcV(pABcyaA), UW551 or ΔHrcV strains (Fig. 1b), indicating that RipAB is secreted by the T3SS, which is consistent with the results of a previous in vitro secretome study (Gueneron et al., 2000; Lonjon et al., 2015).
Figure 1.
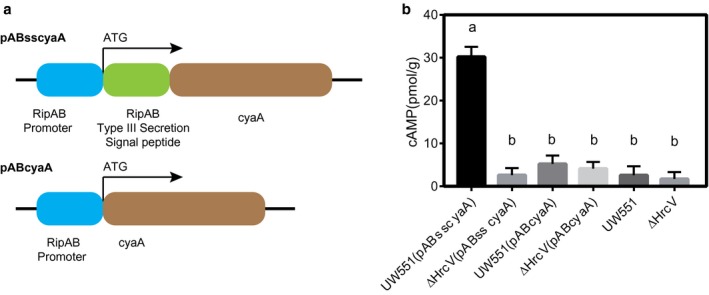
RipAB is a type III‐secreted effector. (a) Schematic representation of the functional domains of the type III secretion system (T3SS) translocation vector pABsscyaA&pABcyaA, including the predicated secretion signal (coding region of 339 bp), the promoter (upstream 200 bp) and cyaA gene. (b) RipAB is translocated into plant cells by a T3SS in vivo. The cyclic adenosine monophosphate (cAMP) levels were measured at 7 days post‐inoculation (dpi). [Means + standard deviations (SDs), n = 3, one‐way analysis of variance (ANOVA) and Tukey’s multiple comparisons test, different letters indicate significant differences, P < 0.01).
RipAB contributes to R. solanacearum UW551 infection in potato
To confirm the virulence screening data, we performed genetic complementation of the ΔripAB mutant. The ΔripAB:ripAB complementation strain was constructed via allelic exchange, in which ripAB expression is driven from its native promoter, and a kanamycin resistance (Kmr) cassette was inserted into a permissive chromosomal site, as described previously (Monteiro et al., 2012). Consistent with previous screening data, potato infection assays showed that the virulence of the ΔripAB strain was significantly reduced (P < 0.0001), whereas the ΔripAB:ripAB complementation strain recovered the virulence to WT levels (Fig. 2a,b).
Figure 2.
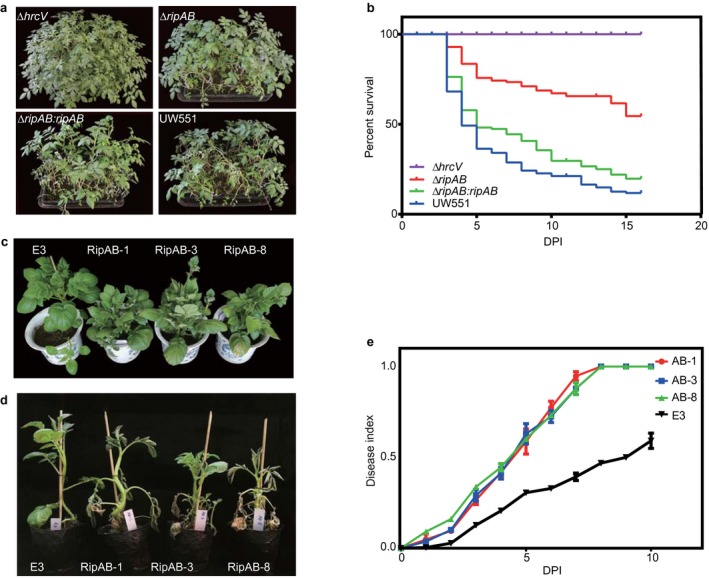
RipAB is required for full virulence in potato. (a) Representative photographs showing the disease symptoms of potato infected by an hrcV mutant, a ripAB mutant, the complementation strain ΔripAB:ripAB and the wild‐type (WT) strain UW551. The potato root‐drenched infections were performed in the glasshouse to evaluate the virulence of effector mutants. The images shown were obtained at 14 days post‐inoculation (dpi). (b) Survival analysis of potato plants inoculated with Ralstonia solanacearum hrcV mutant (purple), ripAB mutant (red), complementation strain ΔripAB:ripAB (green) and WT strain UW551 (blue). One hundred potato plants were inoculated with each individual strain. Statistical analysis was performed via the log‐rank test and the Gehan–Breslow–Wilcoxon test (P < 0.01). (c) Representative photographs of RipAB transgenic lines (RipAB‐1, RipAB‐3 and RipAB‐8). The images shown were obtained 6 weeks after planting. (d) Representative images showing the phenotypes of RipAB transgenic potato lines challenged with WT UW551. The images shown were obtained at 14 dpi. (e) Disease indices of ripAB transgenic potato lines challenged with WT UW551; included are three transgenic lines, RipAB‐1 (red), RipAB‐3 (blue), RipAB‐8 (green), and potato cv. E3 control (black). Each point represents the disease index of three independent experiments, each comprising 24 plants per treatment.
To explore this phenomenon further in potato plants, we generated transgenic potato lines expressing ripAB from a constitutive 35S promoter. In comparison with other transgenic lines expressing control genes under a 35S promoter, all ripAB transgenic lines presented much lower expression of the transgene (Fig. S10, see Supporting Information), suggesting that the plants with high ripAB expression were not viable on genetic transformation with the ripAB expression construct. ripAB transgenic plants exhibited stunted phenotypes (Fig. 2c), suggesting that RipAB alters important plant physiological functions.
Three independent potato transgenic lines (RipAB‐1, RipAB‐3 and RipAB‐8) were subsequently selected and challenged with UW551 (Fig. S11, see Supporting Information). As depicted in Fig. 2d, compared with the untransformed potato cv. E3 control lines, the transgenic lines did not survive for more than 10 dpi with UW551. The three transgenic lines exhibited enhanced disease development, as measured by the disease index (Fig. 2e). Together, these results further confirm that RipAB contributes to UW551 infection in potato.
RipAB requires a nuclear localization signal (NLS) to trigger cell death in N. benthamiana
To predict the molecular function of RipAB, its protein sequence was used to query several protein databases, including the National Center for Biotechnology Information (NCBI), UNIPROT, PHYRE2 and HHpred. However, RipAB did not show any sequence or structural similarity to known proteins. Nevertheless, a nuclear localization signal (NLS) was found in RipAB (Fig. S12, see Supporting Information). To further investigate the function of RipAB, its subcellular localization in plant cells was examined in N. benthamiana using confocal microscopy. A construct with enhanced green fluorescent protein (EGFP) fused to the N‐terminus of RipAB was expressed in N. benthamiana via Agrobacterium‐mediated transient expression. EGFP‐RipAB was observed to localize in the nucleoplasm (Fig. 3a). To determine the relevance of the predicted NLS for RipAB nuclear localization, we generated a RipAB mutant variant with a deletion of the NLS (Fig. S13, see Supporting Information). EGFP‐RipABΔNLS lost the specific nuclear localization (Fig. 3a), indicating that RipAB requires this NLS for its nuclear localization.
Figure 3.
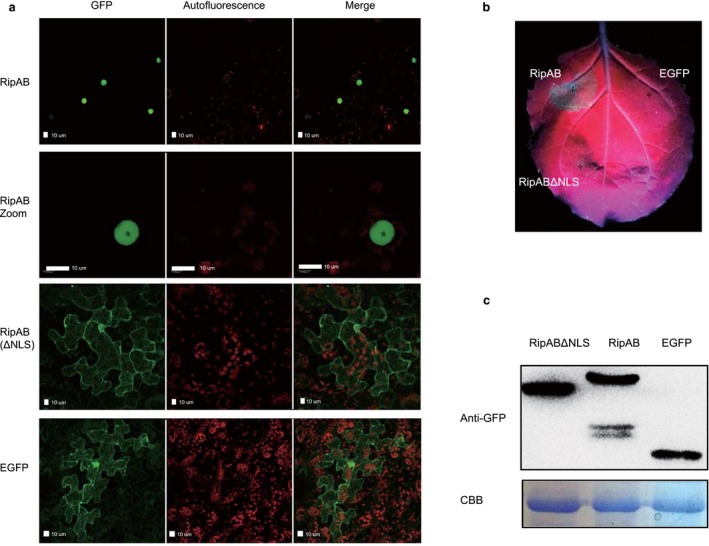
RipAB requires a nuclear localization signal (NLS) to trigger cell death in Nicotiana benthamiana. (a) RipAB localized in the nucleoplasm. The confocal images were captured with a Leica SP8 instrument at 36 h after GV3101 (harbouring vector pk7wg2‐RipAB \pk7wg2‐RipABΔNLS\ pk7wg2)‐mediated transient expression in six‐leaf‐stage N. benthamiana plants. The white scale bar indicates 10 μm. (b) Mutation of NLS abolished the cell death triggered by RipAB in N. benthamiana. pk7wg2 expression plasmids (pk7wg2‐RipAB \pk7wg2‐RipABΔNLS\ pk7wg2) were transformed into Agrobacterium GV3101 for transient expression in N. benthamiana, and the images shown were obtained at 96 h post‐inoculation (hpi). The experiment was conducted in triplicate. (c) Immunoblots of protein extracts from agroinfiltrated leaves of EGFP‐RipAB, EGFP‐RipAB ΔNLS and enhanced green fluorescent protein (EGFP). The proteins were extracted at 48 hpi via extraction buffer with 1% NP40. GFP antibodies were purchased from Thermo Fisher Scientific. The experiment was conducted in triplicate. The protein loading is indicated by Coomassie Brilliant Blue (CBB) staining.
As RipAB triggers cell death in N. benthamiana, we tested whether the NLS is required for cell death induction. EGFP‐RipAB and EGFP‐RipABΔNLS were transiently expressed in N. benthamiana and, as depicted in Fig. 3b,c, EGFP‐RipABΔNLS did not trigger cell death, indicating that the NLS and the nuclear localization of this effector are essential for the induction of cell death.
RipAB suppresses transcriptional responses associated with calcium signalling
To identify specific plant processes affected by RipAB, we performed a transcriptomic analysis of potato plants expressing ripAB. Non‐inoculated roots of three different transgenic lines (RipAB‐1, RipAB‐3 and RipAB‐8) and a control cultivar (E3) were sampled and subjected to RNA sequencing (RNA‐seq) analysis. Four hundred and seventeen differentially expressed genes (DEGs) were identified in all three ripAB transgenic lines compared with the E3 control line (Fig. 4a). Notably, most DEGs were down‐regulated (388), whereas only 29 genes were up‐regulated, which indicates that RipAB interferes with the transcription process (Fig. S14, see Supporting Information).
Figure 4.
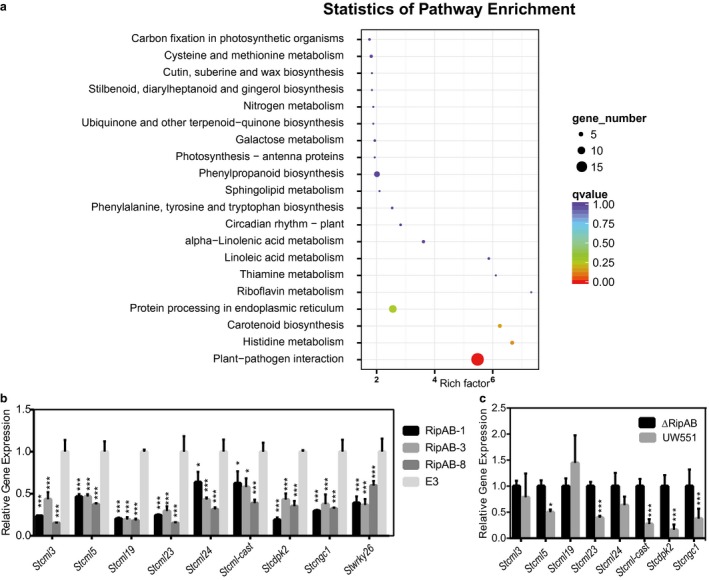
RipAB down‐regulates the calcium signalling pathway at the transcriptional level. (a) Statistics of KEGG pathway enrichment showing that the Plant–Pathogen Interaction (PPI) pathway was the only significantly enriched pathway. Sixteen genes were enriched in the PPI pathway. (b) The relative expression of nine genes involved in the Ca2+ pathway was validated by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) in transgenic plants. (c) The relative expression of eight genes involved in the Ca2+ pathway was validated by qRT‐PCR in potatoes inoculated with mutant UW551 (ΔripAB) and UW551 strains at 24 h post‐inoculation (hpi).
To determine the signalling pathways in which these DEGs are involved, KEGG pathway enrichment analysis was performed. Interestingly, only the Plant Pathogen Interaction (PPI) gene ontology (GO) term was significantly enriched (Fig. 4b). Among all the DEGs, 16 genes were enriched in the PPI GO term, half of which were documented to be involved in Ca2+ signalling, including one cyclic nucleotide‐gated ion channel 1 (Stcngc1) gene, whose product is a calcium transport channel (Jammes et al., 2011), six calmodulin genes, annotated as Stcml3, Stcml5, Stcml19, Stcml23, Stcml24 and Stcml-cast, and one calcium‐dependent protein kinase 2 gene (Stcdpk2), whose product is considered to be a calcium sensor that translates calcium signals into phosphorylation signals (Boudsocq and Sheen, 2013). Additional targeted analysis by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) confirmed that eight calcium signalling‐related genes were significantly down‐regulated in the three ripAB‐expressing transgenic lines (Fig. 4c).
To confirm whether RipAB down‐regulates calcium signalling‐related genes during infection in potato, we investigated gene expression in potato roots after inoculation with UW551 and ΔripAB at 24 hpi. Compared with ΔripAB, infection with UW551 caused a significant down‐regulation of the expression of Stcml5, Stcml23, Stcml‐cast, Stcdpk2 and Stcngc1 (Fig. 4d). These data are consistent with the RNA‐seq data from ripAB transgenic potato plants, strengthening our conclusion that RipAB specifically and significantly down‐regulates calcium signalling‐related genes during infection.
RipAB suppresses the pathogen‐associated molecular pattern (PAMP)‐triggered oxidative burst
As a secondary messenger, Ca2+ is required for PAMP‐induced reactive oxygen species (ROS) production mediated by respiratory burst oxidase homologues (RBOHs) (Kadota et al., 2015). As such, we speculated that the down‐regulation of genes involved in Ca2+‐mediated signal transduction could possibly affect the oxidative burst. Thus, we tested the oxidative burst in transgenic leaf tissues on treatment with the bacterial PAMP flg22, which is the major epitope peptide of bacterial flagellin. In potato plants, the production of ROS reached a peak at 10 min after flg22 treatment. In control plants (E3), a peak value greater than 1000 relative luminescence units (RLU) was reached, whereas the peak value in ripAB‐expressing transgenic lines was less than 500 RLU (Fig. 5a). Compared with the E3 control lines, the ripAB‐expressing transgenic lines exhibited a strong reduction in total ROS generated during 1 h after flg22 treatment (Fig. 5b), suggesting that the down‐regulation of the Ca2+ signalling pathway by RipAB interferes with PAMP‐triggered ROS accumulation.
Figure 5.
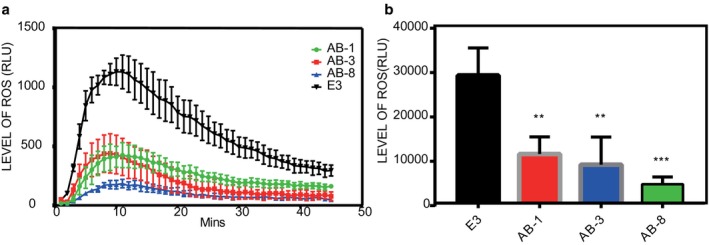
RipAB suppresses the pathogen‐associated molecular pattern (PAMP)‐triggered accumulation of reactive oxygen species (ROS). (a) Time course curve of ROS production in potato leaves, including the three transgenic lines, RipAB‐1 (green), RipAB‐3 (red) and RipAB‐8 (blue), and the potato cultivar E3 control line (black). Each point represents the relative luminescence units (RLU) (mean of three independent experiments), each comprising 24 leaf discs per treatment. The leaf discs were treated with the elicitor flg22 at 100 µm, and flg22‐triggered ROS production was measured as photon counts during a 45‐min period. (b) ROS accumulation during 45 min in potato leaves, including three transgenic lines, RipAB‐1 (red), RipAB‐3 (blue) and RipAB‐8 (green), and the potato cultivar E3 control line (black). [Mean + standard deviation (SD), n = 3, ***P < 0.01, one‐way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test.]
Discussion
Ralstonia solanacearum can infect more than 200 different plant species, and each individual R. solanacearum strain harbours a large effector inventory (60–70) (Genin and Denny, 2012). Thus, R. solanacearum might require different combinations of effectors to infect different hosts. Two lines of evidence exist to support this presumption. First, previous studies have shown that a single effector mutant hardly exhibits any virulence reduction in a limited number of hosts (Macho et al., 2010; Poueymiro et al., 2014). Second, previous studies have shown that R. solanacearum RipAY, a glutamyl cyclotransferase, is the only effector that can significantly reduce glutathione in eggplants, whereas the ripAY mutant shows no marked difference in virulence in eggplants (Fujiwara et al., 2016). This indicates that RipAY, a non‐redundant effector, is dispensable for virulence on eggplants. In this scenario, the virulence screening of effector mutants is necessary for specific host interaction studies and to prioritize effector studies in a large inventory. To investigate which effectors are important for R. solanacearum potato infection, we set up a screen to identify effector mutants with reduced virulence in potato plants. We tested 18 effector mutants, and four exhibited significantly reduced virulence (Fig. S3). Our data suggest that these four effectors contribute to potato infection. However, we must consider that only one accession of susceptible potato was used for this screen, and therefore other relevant effectors could have been overlooked, such as RipAJ and RipG7, which have been shown recently to be under diversifying selection in R. solanacearum (Castillo and Agathos, 2018). In the future, additional accessions will be tested in similar screening studies.
Heterologous expression in yeast is a powerful method to identify conserved biochemical functions in effectors that interfere with yeast growth (Popa et al., 2016). Via this model system, RipAY was revealed as a glutathione‐degrading enzyme which functions in both yeast and plant cells (Fujiwara et al., 2016; Mukaihara et al., 2016; Sang et al., 2018). In keeping with these reports, we also found that RipAY from UW551 inhibits yeast growth, validating our screening approach for the identification of UW551 effectors that interfere with important functions in eukaryotic cells. In addition, RipA5 was found to inhibit the conserved TOR signalling pathway in yeast (Popa et al., 2016). To complement our analysis in yeast cells, we performed transient expression in N. benthamiana. We used cell death as a visualized phenotype (Fig. S5).
The development of cell death can be associated with a severe form of immunity, known as the HR. A previous study has shown that manipulation of the Arabidopsis type I metacaspase regulatory module can almost eliminate HR, but this elimination does not lead to increased pathogen proliferation, decoupling cell death from resistance (Coll et al., 2010). These findings indicate that HR‐type cell death could be independent of immunity and could be mediated by an independent cell death pathway (Coll et al., 2011, 2014; Liu and Levine, 2015). Distinguishing between cell death caused by toxicity and HR is difficult. To identify effectors triggering HR, the corresponding resistance genes (R genes) need to be revealed, which, in most cases, requires additional studies and the development of tools in specific host plants. In our research, four effectors caused cell death, which was independent of the usual signalling components required for immunity triggered by INF1. Therefore, we hypothesize that the cell death triggered by these effectors is unlikely to be a result of an HR, and is most likely caused by a toxic effect of their activities in plant cells. First, their conserved effect on both yeast and N. benthamiana suggests that these effectors could have a conserved biochemical function in eukaryotic cells. Another line of evidence is that cell death triggered by these effectors is slower than that triggered by INF1. In our study, we observed that INF1‐triggered cell death became evident at 36–42 hpi, whereas these effectors triggered cell death at 72–96 hpi.
We observed that several effectors, such as RipAB, RipBH and RipV1, caused multiple phenotypes in our screening system, suggesting a potential association of these three phenotypes (Figs [Link], [Link], [Link]), and raising the possibility that these effectors have a conserved biochemical function or target a conserved signalling pathway in different organisms. It is possible that these effectors target basic cellular functions; for example, RipAB seems to target host processes associated with the Ca2+ signalling pathway, which is essential in all types of biological processes (Cheval et al., 2013).
RipAB was previously named popB (Gueneron et al., 2000). However, the function of RipAB is still unknown. Here, we found that RipAB is a nuclear localized protein required for full virulence during potato infection, and our additional molecular analysis showed that RipAB severely down‐regulates Ca2+‐related signalling at the transcriptional level (Fig. 4). Plants use secondary messengers, such as Ca2+, to perceive stimuli and to adapt to dynamic environments. The alteration of Ca2+ levels can translate perceived signals to downstream biological processes (Cheval et al., 2013). For instance, plant innate immunity requires Ca2+ for the amplification of immune responses (Kadota et al., 2015). Cytosolic Ca2+ concentration dramatically increases after the perception of PAMPs (Lecourieux et al., 2006), resulting in the Ca2+ binding of NADPH oxidases (RBOHD), which increases ROS production (Oda et al., 2010). Such processes can be completed within 15 min and are amongst the first steps for the activation of PAMP‐triggered immunity (PTI) (Boller and Felix, 2009; Macho and Zipfel, 2014). In keeping with this notion, we found that RipAB down‐regulates the expression of Stcngc1 a Ca2+ transport channel gene, and Stcdpk2/Stcmls, Ca2+ sensor genes, accompanied by a reduction in PAMP‐triggered ROS production in ripAB‐expressing transgenic plants (Figs 4 and 6).
Figure 6.
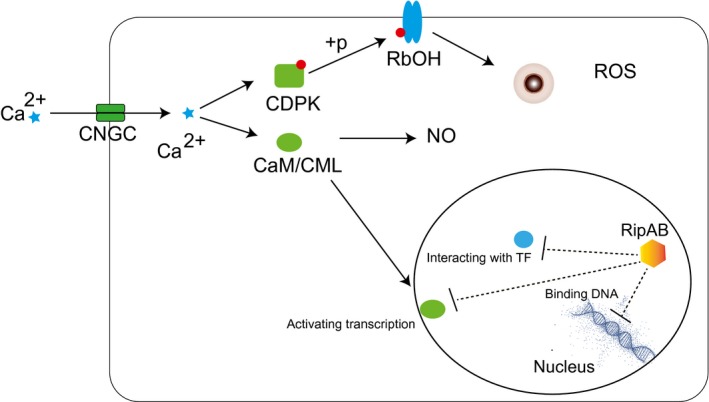
A hypothetical model depicting how RipAB suppresses immunity via the down‐regulation of calcium signalling. RipAB could bind directly to the DNA to impede transcription or could interact with Ca2+ signalling regulators or transcription factors (TF). Down‐regulation of the Ca2+ transport channel gene CNGC would affect Ca2+ influx, and down‐regulation of the calcium sensor would result in the inability of Ca2+ perception, which would lead to the inactivation of downstream immune responses. CaM/CML, Calmodulin; CDPK, calcium‐dependent protein kinase; CNGC, cyclic nucleotide‐gated ion channel; NO, nitric oxide; RBOH, respiratory burst oxidase homologue; ROS, reactive oxygen species.
Notably, a previous study has shown that CDPK/CMLs, Ca2+ sensors, are essential regulators of plant immunity. Mutation of cml24 impairs HR mediated by avrRpt2 (Delk et al., 2005). In addition, silencing of cam13 increases the susceptibility to Tobacco mosaic virus (TMV) and R. solanacearum (Takabatake et al., 2007). Overexpression of cdpk4/5 in potato can phosphorylate RBOHB, leading to HR‐like cell death (Kobayashi et al., 2007). NtCDPK2 autophosphorylation and phosphorylation by an upstream kinase are required for the fine‐tuning of defence signalling cascades (Witte et al., 2010). Ca2+ is a versatile secondary messenger involved in various developmental and adaptive processes in response to various physiological stimuli (Cheval et al., 2013). Therefore, it is important to consider that down‐regulated Ca2+ signalling will not only affect immunity, but also globally alter physiological processes, including hormone signalling, metabolism and responses to other environmental stresses, increasing the vulnerability of the plant to the environment.
We propose two hypothetical scenarios for the mode of action of RipAB (Fig. 6). First, RipAB could bind DNA directly to suppress gene expression associated with Ca2+ signalling. RipAB is a nuclear protein and requires its NLS to trigger cell death, which suggests that RipAB needs to be localized to the nucleus to function properly. Our RNA‐seq data showed that most DEGs were down‐regulated genes (388), which would support a mechanism whereby RipAB directly binds to DNA to impede their transcription. Because RipAB could specifically suppress the Ca2+ signalling pathway at the mRNA level, we speculated that RipAB might mimic a calcium‐related transcription factor to target certain DNA regions. However, the DNA sequence analysis of these down‐regulated calcium‐related genes did not show a clear pattern associated with specific transcription factors. In an alternative scenario, RipAB could interact with Ca2+ signalling regulators, such as calmodulins, and this would result in indirect down‐regulation of the DEGs. Both scenarios would lead to the down‐regulation of the Ca2+ transport channel gene CNGC, which would affect Ca2+ influx. Moreover, a reduced expression of calcium sensors would result in the inability of Ca2+ perception, which would lead to the inactivation of downstream immune responses (Fig. 6).
Experimental Procedures
Strains and vectors
All the strains and vectors used in this study are listed in Table S1 (see Supporting Information).
Plant materials
Potato plants were grown at 18–25 °C in 24‐cm‐diameter plastic pots in the glasshouse (light intensity ranged from 400 to 1000 µE/m2) at the National Centre for Vegetable Improvement (Central China), Huazhong Agricultural University, Wuhan, China.
Solanum tuberosum cv. E‐Potato 3 (E3), susceptible to R. solanacearum, was used for transgenic manipulation, and Solanum chacoense accession C9701 (C9701) was a susceptible wild species used for virulence screening.
Nicotiana benthamiana plants were grown in general‐purpose compost under long‐day conditions of 16 h of light at 22 °C in a glasshouse, and the light intensity ranged from 130 to 150 µE/m2.
Yeast manipulation
Yeast cells were cultured in YPD medium (1% yeast extract, 2% peptone, 2% glucose), and yeast transformations were performed via the lithium acetate method (Gietz and Woods, 1998). Thirty‐three effector genes driven by the galactose promoter were inserted into a pYES2 plasmid for expression. The yeast cells were then cultured for 2 days in synthetic dextrose medium (2% glucose, 0.67% yeast nitrogen base without amino acids) + 2% raffinose, after which the cells were diluted to an optical density at 600 nm (OD600) of 0.4 in water and plated in repression medium (2% glucose) and induction medium (2% galactose) to monitor cell growth. For yeast protein extraction, the yeast cells were grown for 2 days in SD‐Ura medium + 2% raffinose, followed by dilution to an OD600 of 0.4 in liquid induction medium (2% galactose). The cells were subsequently cultured for 12 h before cell lysis.
Transient expression in N. benthamiana
Four‐ to six‐week‐old plants were used for transient expression assays. Agrobacterium tumefaciens cultures (strain GV3101) carrying various constructs were cultured overnight in the presence of the appropriate antibiotics at 28 °C. The bacteria were pelleted, centrifuged and resuspended in infiltration buffer [10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES; pH 5), 10 mm MgCl2 and 0.2 mm acetosyringone], after which their concentration was adjusted to the required OD600 of 0.3 for immunoblots and HR imaging, or 0.05 for confocal imaging. After 3 h of incubation, the cultures were infiltrated into the leaves of N. benthamiana plants. The HR phenotype of the effectors was captured at 60 hpi.
Mutagenesis of R. solanacearum
To generate mutants of R. solanacearum, the target effector genes were replaced with cassettes harbouring Smr. Each sequence included the target gene, and its flanking region amplified from the genome of strain UW551 was inserted into a pEASY‐blunt vector. Reverse amplification of the target gene was used to delete the target gene that generated a linear vector only with the flanking region. An Smr cassette that contained an independent promoter and terminator was inserted into the linear vector using a ClonExpress II One Step Cloning Kit (Vazyme Biotech, Nanjing, China) to create a pEASY‐target::Smr vector. The Smr cassette fragment harbouring the mutant gene flanking region fragments was then amplified and purified for transformation of the R. solanacearum strain, as described previously (Jacobs et al., 2013). All the mutations were verified by both PCR and RT‐PCR, and all the primers used are listed in Table S2 (see Supporting Information). The complementation strain was constructed via allelic exchange, and ripAB with a native promoter and a Kmr cassette were inserted into a permissive chromosomal site, as described previously (Monteiro et al., 2012).
Bacterial motility and growth assays
Bacterial motility assays were carried out on semisolid medium containing 1% tryptone and 0.325 g of agar (Boles and McCarter, 2000). Each plate medium was inoculated with 5 µL of bacterial culture at a density of 107 colony‐forming units (CFU)/mL. The radius of cell movement was measured at 3 dpi. The growth assay was conducted with B medium (Hendrick and Sequeira, 1984) that lacked antibiotics. The cultures were inoculated in 50‐mL tubes containing 15 mL of medium to an initial OD600 of 0.01, and incubated in a 200‐rpm shaker at 28 °C; bacterial growth was measured spectrophotometrically at OD600.
cAMP assays
The cyaA assay was performed as described previously, with some modifications (Poueymiro et al., 2014). Five microlitres of bacterial culture at a density of 107 CFU/mL of strain UW551 and its derivatives harbouring pABsscyaA or pABcyaA plasmids were infiltrated into cut surfaces of potato tubers. The tissues were sampled at 7 days after injection. cAMP levels were monitored with a cAMP enzyme immunoassay kit (New East Biosciences, Malvern, PA, USA) according to the manufacturer’s instructions.
Ralstonia solanacearum infection assays in potato
Two different infection methods were applied in this research.
In vitro potato tests were used for mutant screening as described by Chen et al. (2013), with some modifications. The concentration of inoculated bacteria was adjusted to 107 CFU/mL. Four replicates of 20 plants were investigated for their disease index at 14 dpi. Accession C9701, used for the virulence assays in vitro, was cultivated 3 weeks before inoculation. The disease index was recorded each day after inoculation.
The soil drench method was used for the transgenic lines and complementation virulence assays. With respect to the infection of the transgenic cultivar E3, 24 plants were used for the investigation of the disease index; three replicates were tested. The disease index was recorded each day after inoculation. For the effector complementation virulence assays, 100 plants were used for the investigation of the survival rates. The surviving plants were recorded each day after inoculation, and the concentration of inoculated bacteria was adjusted to 107 CFU/mL.
Confocal laser scanning microscopy
Confocal images of leaf cells expressing the protein fusions at 36 h after transient expression in six‐leaf‐stage N. benthamiana plants were captured with a Leica SP8 (Leica, Wetzlar, Germany) instrument. A wavelength of 488 nm was used for GFP excitation, and the wavelength (500–525 nm) of the emission signal was obtained for the GFP channel. A wavelength of 558 nm was used for chloroplast fluorescence excitation, and emission wavelengths (690–720 nm) were obtained.
Immunoblot analysis
Samples were collected and ground in liquid nitrogen. Lysis buffer was then used as described previously (Kadota et al., 2016). The proteins were separated by 10% sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE), transferred to a polyvinylidene fluoride (PVDF) membrane (Biorad, Hercules, CA, USA) and then measured via an ECL plus (Biorad) device. The anti‐GFP and horseradish peroxidase‐conjugated secondary antibodies used in this research were purchased from Thermo Fisher Scientific, Shanghai, China.
Potato genetic transformation
The ripAB gene driven by P35S was transformed into potato cultivar E3 via A. tumefaciens (LBA4404)‐mediated transformation, as described previously (Tian et al., 2003). The regenerated plants were identified by semiquantitative PCR.
Transcriptome analysis
In this study, root samples of 4‐week‐old in vitro potatoes were harvested. Taking untransformed potato cultivar E3 as a control, we sampled three ripAB transgenic lines to constitute three biological replicates. The total RNA was extracted with an RNA extraction kit (Zoman Biotechnology, Beijing, China) according to the manufacturer’s instructions. RNA‐seq was performed by Biomarker Technologies (Beijing, China) with a HiSeq‐xTEN (Illumina, San Diego, CA, USA) system. The data were analysed by the BMK Cloud server. The gene expression levels were quantified by the fragments per kilobase of transcript per million fragments mapped (FPKM) method (Trapnell et al., 2010). A value of |log2(fold change)| ≥ 1 [false discovery rate (FDR) < 0.01] was set as the threshold for DEGs. GO enrichment analysis of the DEGs was implemented by the GOseq R package (Ashburner et al., 2000), and the enrichment of the DEGs in the KEGG pathways was analysed via KOBAS software (Mao et al., 2005). Raw sequencing data are available in the Sequence Read Archive under the accession code: PRJNA500722.
qRT‐PCR
Total RNA was extracted using a Plant RNA kit (Biotek, Winooski, VT, USA) in accordance with the manufacturer’s instructions. First‐strand cDNA was synthesized using an iScript cDNA synthesis kit (Biorad). qRT‐PCR was performed using SYBR Green Supermix (Biorad) and a CFX96 Real Time system (Biorad), and the qRT‐PCR data were analysed as described previously (Livak and Schmittgen, 2001). The potato gene ef1α (GenBank accession: AB061263) was used as an internal reference gene (Nicot et al., 2005). The primer sets used for qRT‐PCR analysis are listed in Table S2.
ROS assays
Oxidative bursts were measured as described previously (Gimenez‐Ibanez et al., 2009; Sang and Macho, 2017), with some modifications. ROS were elicited with 100 nm flg22, and luminescence was measured during a 45‐min period using a microplate luminescence reader. The luminescence was recorded in each well for 400 ms each minute. The data obtained from the ROS burst assays were analysed and represented in two different ways: the RLU produced per minute (the kinetics of ROS production) and the total cumulative values of RLU for each sample (the total ROS production).
VIGS assays
VIGS assays were performed as described by Liu et al. (2005). The primers used are listed in Table S2.
Supporting information
Fig. S1 Bacterial motility of effector mutants. Bacterial motility was assayed on semisolid medium. Each plate medium was inoculated with 5 µL of bacterial culture at a density of 107 colony‐forming units (CFU)/mL. The radius of cell movement was collected at 3 days post‐inoculation (dpi). [Mean + standard deviation (SD), n = 3, ***P < 0.01, one‐way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test.]
Fig. S2 Bacterial growth of effector mutants. Bacterial growth was assayed with B medium (Hendrick and Sequeira, 1984) that lacked antibiotics. The optical density at 600 nm were monitored at 2 days post‐inoculation (dpi). [Mean + standard deviation (SD), n = 3, ***P < 0.01, one‐way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test.]
Fig. S3 Virulence screening of Ralstonia solanacearum UW551 effector mutants in potato. The potato unwounded root infection in vitro test was used, and the disease grade was determined at 14 days post‐inoculation (dpi). Twenty‐four plants per test were inoculated with strains at a density of 107 colony‐forming units (CFU)/mL. [Mean + standard deviation (SD), n = 4, ***P < 0.01, one‐way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test.]
Fig. S4 Bacterial growth curve of effector mutants. Bacterial growth was assayed with B medium (Hendrick and Sequeira, 1984) that lacked antibiotics. The (optical density at 600 nm) of ΔripAB, ΔripV1, ΔripBH, ΔripF2 and UW551 were monitored spectrophotometrically during a 60‐h period.
Fig. S5 Ralstonia solanacearum UW551 effectors interfere with yeast growth under two conditions. PYES‐nta plasmids fused with effector proteins were transformed into yeast BY4741 and then subjected to synthetic dextrose medium comprising different carbon sources, such as galactose (induction) or glucose (suppression). The yeast growth inhibition screening was performed under two conditions (normal conditions and salt stress conditions with 0.5 m NaCl).
Fig. S6 Representative graphs of four effectors developing a cell death phenotype in Nicotiana benthamiana. The expression plasmids PH7C10.0 with effectors were transformed into Agrobacterium GV3101 for transient expression in N. benthamiana, and images were taken at 96 h post‐inoculation (hpi) (n = 4).
Fig. S7 Average inoculations causing the development of cell death phenotypes for RipAB, RipBH and RipV1. Thirty inoculations were performed with individual effectors. The number of dead cells was counted at 96 h post‐inoculation (hpi). [Mean + standard deviation (SD), n = 3, letters indicate significant differences P < 0.01, one‐way analysis of variance (ANOVA) and Tukey’s multiple comparisons test.]
Fig. S8 Average inoculations causing the development of cell death phenotypes for RipAB, RipBH and RipV1 with respect to virus‐induced gene silencing (VIGS) of plant immune response‐related genes (NbSGT1, NbEDS1, bNDR1, NbHSP70 and NbMEKK2). Agroinfiltration was performed with Nicotiana benthamiana plants at 3 weeks after gene silencing. Thirty inoculations were performed with individual effectors in each gene‐silenced plant. The number of dead cells was counted at 96 h post‐inoculation (hpi). [Mean + standard deviation (SD), n = 4, ***P < 0.001, one‐way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test.]
Fig. S9 Relative gene expression in plants subjected to virus‐induced gene silencing (VIGS). Plant leaves were sampled at 3 weeks after VIGS. [Means + standard deviations (SDs), n = 3, ***P < 0.01, one‐way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test.]
Fig. S10 Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of RipAB and GUS transgenic potato plants. Four‐week‐old potato leaves were sampled. [Means + standard deviations (SDs), n = 3].
Fig. S11 Semi‐quantitative polymerase chain reaction (PCR) of regenerated RipAB transgenic potato plants. Seven transgenic plants of RipAB were sampled and subjected to semiquantative PCR.
Fig. S12 Prediction of nuclear localization signals (NLSs). The green letters indicate NLSs.
Fig. S13 Schematic representation of the nuclear localization signal (NLS) of RipAB.
Fig. S14 Heat map showing the expression patterns of RipAB transgenic lines and cv. E3. The hierarchical clustering is shown of 417 genes identified as differentially expressed in the comparison between RipAB transgenic lines and cv. E3.
Table S1 Strains and plasmids.
Table S2 List of primers used for quantitative polymerase chain reaction (qPCR) analysis and mutant construction.
Acknowledgements
We are grateful for the useful advice on this project from Feng Li and Shunping Yan. We thank Zhendong Tian, Qin He, Juan Du and Bo Li for critical reading and suggestions on the manuscript. We thank Caitilyn Allen and Philippe Prior for providing us with R. solanacearum strains. We also thank all members of the potato group in Huazhong Agricultural University for supporting this project. This project was supported by the Fundamental Research Funds for the Central Universities (Grant No. 2013PY080) and the Initial Project Fund from Huazhong Agricultural University (Grant No. 12012). A.P.M. is supported by the Shanghai Center for Plant Stress Biology (Chinese Academy of Sciences) and the Chinese 1000 Talents Program. Botao Song is supported by the Earmarked Fund for Modern Agro‐Industry Technology Research System of China (CARS‐09‐P07). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- Angot, A. , Peeters, N. , Lechner, E. , Vailleau, F. , Baud, C. , Gentzbittel, L. , Sartorel, E. , Genschik, P. , Boucher, C. and Genin, S. (2006) Ralstonia solanacearum requires F‐box‐like domain‐containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. USA, 103, 14 620–14 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M. , Ball, C.A. , Blake, J.A. , Botstein, D. , Butler, H. , Cherry, J.M. , Davis, A.P. , Dolinski, K. , Dwight, S.S. and Eppig, J.T. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block, A. and Alfano, J.R. (2011) Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr. Opin. Microbiol. 14, 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles, B.R. and McCarter, L.L. (2000) Insertional inactivation of genes encoding components of the sodium‐type flagellar motor and switch of Vibrio parahaemolyticus. J. Bacteriol. 182, 1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Bombarely, A. , Rosli, H.G. , Vrebalov, J. , Moffett, P. , Mueller, L.A. and Martin, G.B. (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant‐microbe biology research. Mol. Plant–Microbe Interact. 25, 1523. [DOI] [PubMed] [Google Scholar]
- Bosis, E. , Salomon, D. and Sessa, G. (2011) A simple yeast‐based strategy to identify host cellular processes targeted by bacterial effector proteins. Plos One, 6, e27698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq, M. and Sheen, J. (2013) CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein, D. , Zusman, T. , Degtyar, E. , Viner, R. , Segal, G. and Pupko, T. (2009) Genome‐scale identification of Legionella pneumophila effectors using a machine learning approach. Plos Pathogens, 5, e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo, J.A. and Agathos, S.N. (2018). Candidate genes under balancing selection in a plant bacterial pathogen. bioRxiv , 388207.
- Champoiseau, P.G. , Jones, J.B. and Allen, C. (2009) Ralstonia solanacearum Race 3 biovar 2 causes tropical losses and temperate anxieties. Plant Health Progress, 10, 1–10. [Google Scholar]
- Chen, L. , Guo, X. , Wang, H. , Xie, C. , Cai, X. , He, L. , Zhou, J. and Liu, J. (2016) Tetrasomic inheritance pattern of the pentaploid Solanum chacoense (+) S. tuberosum somatic hybrid (resistant to bacterial wilt) revealed by SSR detected alleles. Plant Cell, Tissue and Organ Culture (PCTOC), 127, 1–9. [Google Scholar]
- Chen, L. , Guo, X. , Xie, C. , He, L. , Cai, X. , Tian, L. , Song, B. and Liu, J. (2013) Nuclear and cytoplasmic genome components of Solanum tuberosum + S. chacoense somatic hybrids and three SSR alleles related to bacterial wilt resistance. Theor. Appl. Genet. 126, 1861–1872. [DOI] [PubMed] [Google Scholar]
- Cheval, C. , Aldon, D. , Galaud, J.P. and Ranty, B. (2013) Calcium/calmodulin‐mediated regulation of plant immunity. Biochim. Biophys. Acta, 1833, 1766–1771. [DOI] [PubMed] [Google Scholar]
- Coll, N.S. , Epple, P. and Dangl, J.L. (2011) Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll, N.S. , Smidler, A. , Puigvert, M. , Popa, C. , Valls, M. and Dangl, J.L. (2014) The plant metacaspase AtMC1 in pathogen‐triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ. 21, 1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll, N.S. , Vercammen, D. , Smidler, A. , Clover, C. , van Breusegem, F. , Dangl, J.L. and Epple, P. (2010) Arabidopsis type I metacaspases control cell death. Science, 330, 1393–1397. [DOI] [PubMed] [Google Scholar]
- Curak, J. , Rohde, J. and Stagljar, I. (2009) Yeast as a tool to study bacterial effectors. Curr. Opin. Microbiol. 12, 18–23. [DOI] [PubMed] [Google Scholar]
- Delk, N.A. , Johnson, K.A. , Chowdhury, N.I. and Braam, J. (2005) CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 139, 240–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L. and Genin, S. (2014) Opening the Ralstonia solanacearum type III effector tool box: insights into host cell subversion mechanisms. Curr. Opin. Plant Biol. 20C, 110–117. [DOI] [PubMed] [Google Scholar]
- Du, J. , Verzaux, E. , Chaparro‐Garcia, A. , Bijsterbosch, G. , Keizer, L.C.P. , Zhou, J. , Liebrand, T.W.H. , Xie, C.H. , Govers, F. , Robatzek, S. , van der Vossen, E.A.G. , Jacobsen, E. , Visser, R.G.F. , Kamoun, S. and Vleeshouwers, V.G.A.A. (2015) Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants, 1, 15034 [DOI] [PubMed] [Google Scholar]
- Fegan, M. , Prior, P. , Allen, C. , Prior, P. and Hayward, A.C. (2005) How complex is the Ralstonia solanacearum species complex? APS press, pp. 449–461. [Google Scholar]
- Fujiwara, S. , Kawazoe, T. , Ohnishi, K. , Kitagawa, T. , Popa, C. , Valls, M. , Genin, S. , Nakamura, K. , Kuramitsu, Y. , Tanaka, N. and Tabuchi, M. (2016) RipAY, a plant pathogen effector protein, exhibits robust γ‐glutamyl cyclotransferase activity when stimulated by eukaryotic thioredoxins. J. Biol. Chem. 291, 6813–6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin, S. and Denny, T.P. (2012) Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D. and Woods, R.A. (1998) 4 transformation of yeast by the lithium acetate/single‐stranded carrier DNA/PEG method. Methods Microbiol. 26, 53–66. [Google Scholar]
- Gimenez‐Ibanez, S. , Hann, D.R. , Ntoukakis, V. , Petutschnig, E. , Lipka, V. and Rathjen, J.P. (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429. [DOI] [PubMed] [Google Scholar]
- Goodin, M.M. , Zaitlin, D. , Naidu, R.A. and Lommel, S.A. (2008) Nicotiana benthamiana: its history and future as a model for plant‐pathogen interactions. Mol. Plant–Microbe Interact. 21, 1015–1026. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. and Vinatzer, B.A. (2003) Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr. Opin. Microbiol. 6, 20–28. [DOI] [PubMed] [Google Scholar]
- Gueneron, M. , Timmers, A.C.J. , Boucher, C. and Arlat, M. (2000) Two novel proteins, PopB, which has functional nuclear localization signals, and PopC, which has a large leucine‐rich repeat domain, are secreted through the Hrp‐secretion apparatus of Ralstonia solanacearum . Mol. Microbiol. 36, 261–277. [DOI] [PubMed] [Google Scholar]
- Hayward, A.C. (1964) Characteristics of Pseudomonas solanacearum . J. Appl. Microbiol. 27, 265–277. [Google Scholar]
- Hayward, A.C. (2003) Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum . Annu. Rev. Phytopathol. 29, 65–87. [DOI] [PubMed] [Google Scholar]
- Hendrick, C.A. and Sequeira, L. (1984) Lipopolysaccharide‐defective mutants of the wilt pathogen Pseudomonas‐solanacearum . Appl. Environ. Microbiol. 48, 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker, W.J. (1981) Compendium of potato diseases International Potato Center. Pp. 1–20. [Google Scholar]
- Huerta, A.I. , Milling, A. and Allen, C. (2015) Tropical strains of Ralstonia solanacearum outcompete Race 3 Biovar 2 strains at lowland tropical temperatures. Appl. Environ. Microbiol. 81, 3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, J.M. , Milling, A. , Mitra, R.M. , Hogan, C.S. , Ailloud, F. , Prior, P. and Allen, C. (2013) Ralstonia solanacearum requires PopS, an ancient AvrE‐family effector, for virulence and to overcome salicylic acid‐mediated defenses during tomato pathogenesis. MBio, 4, e00875–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammes, F. , Hu, H.C. , Villiers, F. , Bouten, R. and Kwak, J.M. (2011) Calcium‐permeable channels in plant cells. FEBS J. 278, 4262–4276. [DOI] [PubMed] [Google Scholar]
- Janse, J.D. (1996) Potato brown rot in western Europe – history, present occurrence and some remarks on possible origin, epidemiology and control strategies. Eppo Bull. 26, 679–695. [Google Scholar]
- Kadota, Y. , Macho, A.P. and Zipfel, C. (2016) Immunoprecipitation of plasma membrane receptor‐like kinases for identification of phosphorylation sites and associated proteins. Methods Mol. Biol. 1363, 133–144. [DOI] [PubMed] [Google Scholar]
- Kadota, Y. , Shirasu, K. and Zipfel, C. (2015) Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56, 1472–1480. [DOI] [PubMed] [Google Scholar]
- Kay, S. and Bonas, U. (2009) How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 12, 37–43. [DOI] [PubMed] [Google Scholar]
- Kobayashi, M. , Ohura, I. , Kawakita, K. , Yokota, N. , Fujiwara, M. , Shimamoto, K. , Doke, N. and Yoshioka, H. (2007) Calcium‐dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell, 19, 1065–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux, C. , Huet, G. , Jauneau, A. , Camborde, L. , Tremousaygue, D. , Kraut, A. , Zhou, B. , Levaillant, M. , Adachi, H. , Yoshioka, H. , Raffaele, S. , Berthome, R. , Coute, Y. , Parker, J.E. and Deslandes, L. (2015) A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell, 161, 1074–1088. [DOI] [PubMed] [Google Scholar]
- Lecourieux, D. , Ranjeva, R. and Pugin, A. (2006) Calcium in plant defence‐signalling pathways. New Phytol. 171, 249–269. [DOI] [PubMed] [Google Scholar]
- Lindeberg, M. , Cunnac, S. and Collmer, A. (2012) Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 20, 199. [DOI] [PubMed] [Google Scholar]
- Liu, Y. and Levine, B. (2015) Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ. 22, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Czymmek, K. , Talloczy, Z. , Levine, B. and Dinesh‐Kumar, S.P. (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell, 121, 567–577. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lonjon, F. , Peeters, N. , Genin, S. and Vailleau, F. (2018) In vitro and in vivo secretion/translocation assays to identify novel Ralstonia solanacearum type 3 effectors In: Host-Pathogen Interactions, Medina C., López-Baena F. (eds), (pp. 209–222). New York: Humana Press. [DOI] [PubMed] [Google Scholar]
- Lonjon, F. , Turner, M. , Henry, C. , Rengel, D. , Lohou, D. , van de Kerkhove, Q. , Cazale, A.C. , Peeters, N. , Genin, S. and Vailleau, F. (2015) Comparative secretome analysis of Ralstonia solanacearum type 3 secretion‐associated mutants reveals a fine control of effector delivery, essential for bacterial pathogenicity. Mol. Cell Proteomics, 15, 598–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho, A.P. (2016) Subversion of plant cellular functions by bacterial type‐III effectors: beyond suppression of immunity. New Phytol, 210, 51–57. [DOI] [PubMed] [Google Scholar]
- Macho, A.P. , Guidot, A. , Barberis, P. , Beuzon, C.R. and Genin, S. (2010) A competitive index assay identifies several Ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants. Mol. Plant–Microbe Interact. 23, 1197–1205. [DOI] [PubMed] [Google Scholar]
- Macho, A.P. and Zipfel, C. (2014) Plant PRRs and the activation of innate immune signaling. Mol. Cell, 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Macho, A.P. and Zipfel, C. (2015) Targeting of plant pattern recognition receptor‐triggered immunity by bacterial type‐III secretion system effectors. Curr. Opin. Microbiol. 23, 14–22. [DOI] [PubMed] [Google Scholar]
- Mao, X. , Cai, T. , Olyarchuk, J.G. and Wei, L. (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics, 21, 3787–3793. [DOI] [PubMed] [Google Scholar]
- McIntyre, D.F. (2003) Food and agriculture organization of the United Nations. Encycl. Food Sci. Nutr. 1, 2587–2593. [Google Scholar]
- Monteiro, F. , Sole, M. , van Dijk, I. and Valls, M. (2012) A chromosomal insertion toolbox for promoter probing, mutant complementation, and pathogenicity studies in Ralstonia solanacearum . Mol. Plant–Microbe Interact. 25, 557–568. [DOI] [PubMed] [Google Scholar]
- Mudgett, M.B. (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56, 509–531. [DOI] [PubMed] [Google Scholar]
- Mukaihara, T. , Hatanaka, T. , Nakano, M. and Oda, K. (2016) Ralstonia solanacearum type III effector RipAY is a glutathione‐degrading enzyme that is activated by plant cytosolic thioredoxins and suppresses plant immunity. Mbio 7, e00359–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot, N. , Hausman, J.F. , Hoffmann, L. and Evers, D. (2005) Reference gene selection for RT‐qPCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 56, 2907–2914. [DOI] [PubMed] [Google Scholar]
- Oda, T. , Hashimoto, H. , Kuwabara, N. , Akashi, S. , Hayashi, K. , Kojima, C. , Wong, H.L. , Kawasaki, T. , Shimamoto, K. and Sato, M. (2010) Structure of the N‐terminal regulatory domain of a plant NADPH oxidase and its functional implications. J. Biol. Chem. 285, 1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S.K. , Young, C. , Lee, M. , Oliva, R. , Bozkurt, T.O. , Cano, L.M. , Win, J. , Bos, J.I. , Liu, H.Y. and Van, D.M. (2009) In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi‐blb2. Plant Cell, 21, 2928–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, N. , Carrere, S. , Anisimova, M. , Plener, L. , Cazale, A.C. and Genin, S. (2013a) Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics, 14, 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, N. , Guidot, A. , Vailleau, F. and Valls, M. (2013b) Ralstonia solanacearum, a widespread bacterial plant pathogen in the post‐genomic era. Mol. Plant Pathol. 14, 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petnickiocwieja, T. , Schneider, D.J. , Tam, V.C. , Chancey, S.T. , Shan, L. , Jamir, Y. , Schechter, L.M. , Janes, M.D. , Buell, C.R. and Tang, X. (2002) Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA, 99 , 7652–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa, C. , Li, L. , Gil, S. , Tatjer, L. , Hashii, K. , Tabuchi, M. , Coll, N.S. , Arino, J. and Valls, M. (2016) The effector AWR5 from the plant pathogen Ralstonia solanacearum is an inhibitor of the TOR signalling pathway. Sci. Rep. 6, 27 058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro, M. , Cazale, A.C. , Francois, J.M. , Parrou, J.L. , Peeters, N. and Genin, S. (2014) A Ralstonia solanacearum type III effector directs the production of the plant signal metabolite trehalose‐6‐phosphate. MBio, 5, e02065–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro, M. , Cunnac, S. , Barberis, P. , Deslandes, L. , Peeters, N. , Cazale‐Noel, A.C. , Boucher, C. and Genin, S. (2009) Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host‐range specificity on tobacco. Mol. Plant–Microbe Interact. 22, 538–550. [DOI] [PubMed] [Google Scholar]
- Puigvert, M. , Guarischi‐Sousa, R. , Zuluaga, P. , Coll, N.S. , Macho, A.P. , Setubal, J.C. and Valls, M. (2017) Transcriptomes of Ralstonia solanacearum during root colonization of Solanum commersonii. Front. Plant Sci. 8, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remigi, P. , Anisimova, M. , Guidot, A. , Genin, S. and Peeters, N. (2011) Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol. 192, 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon, D. , Dar, D. , Sreeramulu, S. and Sessa, G. (2011) Expression of Xanthomonas campestris pv. vesicatoria type III effectors in yeast affects cell growth and viability. Mol. Plant–Microbe Interact. 24, 305. [DOI] [PubMed] [Google Scholar]
- Sang, Y. and Macho, A.P. (2017) Analysis of PAMP‐triggered ROS burst in plant immunity. Methods Mol. Biol. 1578, 143–153. [DOI] [PubMed] [Google Scholar]
- Sang, Y.Y. , Wang, Y.R. , Ni, H. , Cazale, A.C. , She, Y.M. , Peeters, N. and Macho, A.P. (2018) The Ralstonia solanacearum type III effector RipAY targets plant redox regulators to suppress immune responses. Mol. Plant Pathol. 19, 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris, P.F. , Duxbury, Z. , Huh, S.U. , Ma, Y. , Segonzac, C. , Sklenar, J. , Derbyshire, P. , Cevik, V. , Rallapalli, G. , Saucet, S.B. , Wirthmueller, L. , Menke, F.L. , Sohn, K.H. and Jones, J.D. (2015) A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell, 161, 1089–1100. [DOI] [PubMed] [Google Scholar]
- Seal, S.E. , Taghavi, M. , Fegan, N. , Hayward, A.C. and Fegan, M. (1999) Determination of Ralstonia (Pseudomonas) solanacearum rDNA subgroups by PCR tests. Plant Pathol. 48, 115–120. [Google Scholar]
- Sun, Y.H. , Li, P. , Deng, M.Y. , Shen, D. , Dai, G.Y. , Yao, N. and Lu, Y.J. (2017) The Ralstonia solanacearum effector RipAK suppresses plant hypersensitive response by inhibiting the activity of host catalases. Cell. Microbiol. 19, e12736. [DOI] [PubMed] [Google Scholar]
- Takabatake, R. , Karita, E. , Seo, S. , Mitsuhara, I. , Kuchitsu, K. and Ohashi, Y. (2007) Pathogen‐induced calmodulin isoforms in basal resistance against bacterial and fungal pathogens in tobacco. Plant Cell Physiol. 48, 414–423. [DOI] [PubMed] [Google Scholar]
- Tian, Z.D. , Liu, J. and Xie, C.H. (2003) Isolation of resistance‐related genes to Phytophthora infestans with suppression subtractive hybridization in the R‐gene‐free potato. Yi Chuan Xue Bao, 30, 597–605. [PubMed] [Google Scholar]
- Trapnell, C. , Williams, B.A. , Pertea, G. , Mortazavi, A. , Kwan, G. , van Baren, M.J. , Salzberg, S.L. , Wold, B.J. and Pachter, L. (2010) Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon, Z. , White, D. , Jørgensen, R. , Visschedyk, D. , Fieldhouse, R.J., Mangroo, D. and Merrill, A.R. (2009) Yeast as a tool for characterizing mono‐ADP‐ribosyltransferase toxins. Fems Microbiol. Lett. 300, 97–106. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Remigi, P. , Anisimova, M. , Lonjon, F. , Kars, I. , Kajava, A. , Li, C.H. , Cheng, C.P. , Vailleau, F. , Genin, S. and Peeters, N. (2015) Functional assignment to positively selected sites in the core type III effector RipG7 from Ralstonia solanacearum . Mol. Plant Pathol, 17, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Wang, B. , Zhao, G. , Cai, X. , Jabaji, S. , Seguin, P. and Chen, H. (2017) Genetic and pathogenic diversity of Ralstonia solanacearum causing potato brown rot in China. Am. J. Potato Res, 94, 403–416. [Google Scholar]
- Wicker, E. , Lefeuvre, P. , de Cambiaire, J.C. , Lemaire, C. , Poussier, S. and Prior, P. (2012) Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. ISME J. 6, 961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte, C.P. , Keinath, N. , Dubiella, U. , Demouliere, R. , Seal, A. and Romeis, T. (2010) Tobacco calcium‐dependent protein kinases are differentially phosphorylated in vivo as part of a kinase cascade that regulates stress response. J. Biol. Chem. 285, 9740–9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullings, B.A. , Beuningen, A.R.V. , Janse, J.D. and Akkermans, A.D.L. (1998) Detection of Ralstonia solanacearum, which causes brown rot of potato, by fluorescent in situ hybridization with 23S rRNA‐targeted probes. Appl. Environ. Microbiol. 64, 4546–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M. and Chai, J. (2008) Plant pathogenic bacterial type III effectors subdue host responses. Curr. Opin. Microbiol. 11, 179–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Bacterial motility of effector mutants. Bacterial motility was assayed on semisolid medium. Each plate medium was inoculated with 5 µL of bacterial culture at a density of 107 colony‐forming units (CFU)/mL. The radius of cell movement was collected at 3 days post‐inoculation (dpi). [Mean + standard deviation (SD), n = 3, ***P < 0.01, one‐way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test.]
Fig. S2 Bacterial growth of effector mutants. Bacterial growth was assayed with B medium (Hendrick and Sequeira, 1984) that lacked antibiotics. The optical density at 600 nm were monitored at 2 days post‐inoculation (dpi). [Mean + standard deviation (SD), n = 3, ***P < 0.01, one‐way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test.]
Fig. S3 Virulence screening of Ralstonia solanacearum UW551 effector mutants in potato. The potato unwounded root infection in vitro test was used, and the disease grade was determined at 14 days post‐inoculation (dpi). Twenty‐four plants per test were inoculated with strains at a density of 107 colony‐forming units (CFU)/mL. [Mean + standard deviation (SD), n = 4, ***P < 0.01, one‐way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test.]
Fig. S4 Bacterial growth curve of effector mutants. Bacterial growth was assayed with B medium (Hendrick and Sequeira, 1984) that lacked antibiotics. The (optical density at 600 nm) of ΔripAB, ΔripV1, ΔripBH, ΔripF2 and UW551 were monitored spectrophotometrically during a 60‐h period.
Fig. S5 Ralstonia solanacearum UW551 effectors interfere with yeast growth under two conditions. PYES‐nta plasmids fused with effector proteins were transformed into yeast BY4741 and then subjected to synthetic dextrose medium comprising different carbon sources, such as galactose (induction) or glucose (suppression). The yeast growth inhibition screening was performed under two conditions (normal conditions and salt stress conditions with 0.5 m NaCl).
Fig. S6 Representative graphs of four effectors developing a cell death phenotype in Nicotiana benthamiana. The expression plasmids PH7C10.0 with effectors were transformed into Agrobacterium GV3101 for transient expression in N. benthamiana, and images were taken at 96 h post‐inoculation (hpi) (n = 4).
Fig. S7 Average inoculations causing the development of cell death phenotypes for RipAB, RipBH and RipV1. Thirty inoculations were performed with individual effectors. The number of dead cells was counted at 96 h post‐inoculation (hpi). [Mean + standard deviation (SD), n = 3, letters indicate significant differences P < 0.01, one‐way analysis of variance (ANOVA) and Tukey’s multiple comparisons test.]
Fig. S8 Average inoculations causing the development of cell death phenotypes for RipAB, RipBH and RipV1 with respect to virus‐induced gene silencing (VIGS) of plant immune response‐related genes (NbSGT1, NbEDS1, bNDR1, NbHSP70 and NbMEKK2). Agroinfiltration was performed with Nicotiana benthamiana plants at 3 weeks after gene silencing. Thirty inoculations were performed with individual effectors in each gene‐silenced plant. The number of dead cells was counted at 96 h post‐inoculation (hpi). [Mean + standard deviation (SD), n = 4, ***P < 0.001, one‐way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test.]
Fig. S9 Relative gene expression in plants subjected to virus‐induced gene silencing (VIGS). Plant leaves were sampled at 3 weeks after VIGS. [Means + standard deviations (SDs), n = 3, ***P < 0.01, one‐way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test.]
Fig. S10 Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of RipAB and GUS transgenic potato plants. Four‐week‐old potato leaves were sampled. [Means + standard deviations (SDs), n = 3].
Fig. S11 Semi‐quantitative polymerase chain reaction (PCR) of regenerated RipAB transgenic potato plants. Seven transgenic plants of RipAB were sampled and subjected to semiquantative PCR.
Fig. S12 Prediction of nuclear localization signals (NLSs). The green letters indicate NLSs.
Fig. S13 Schematic representation of the nuclear localization signal (NLS) of RipAB.
Fig. S14 Heat map showing the expression patterns of RipAB transgenic lines and cv. E3. The hierarchical clustering is shown of 417 genes identified as differentially expressed in the comparison between RipAB transgenic lines and cv. E3.
Table S1 Strains and plasmids.
Table S2 List of primers used for quantitative polymerase chain reaction (qPCR) analysis and mutant construction.


