Figure 1.
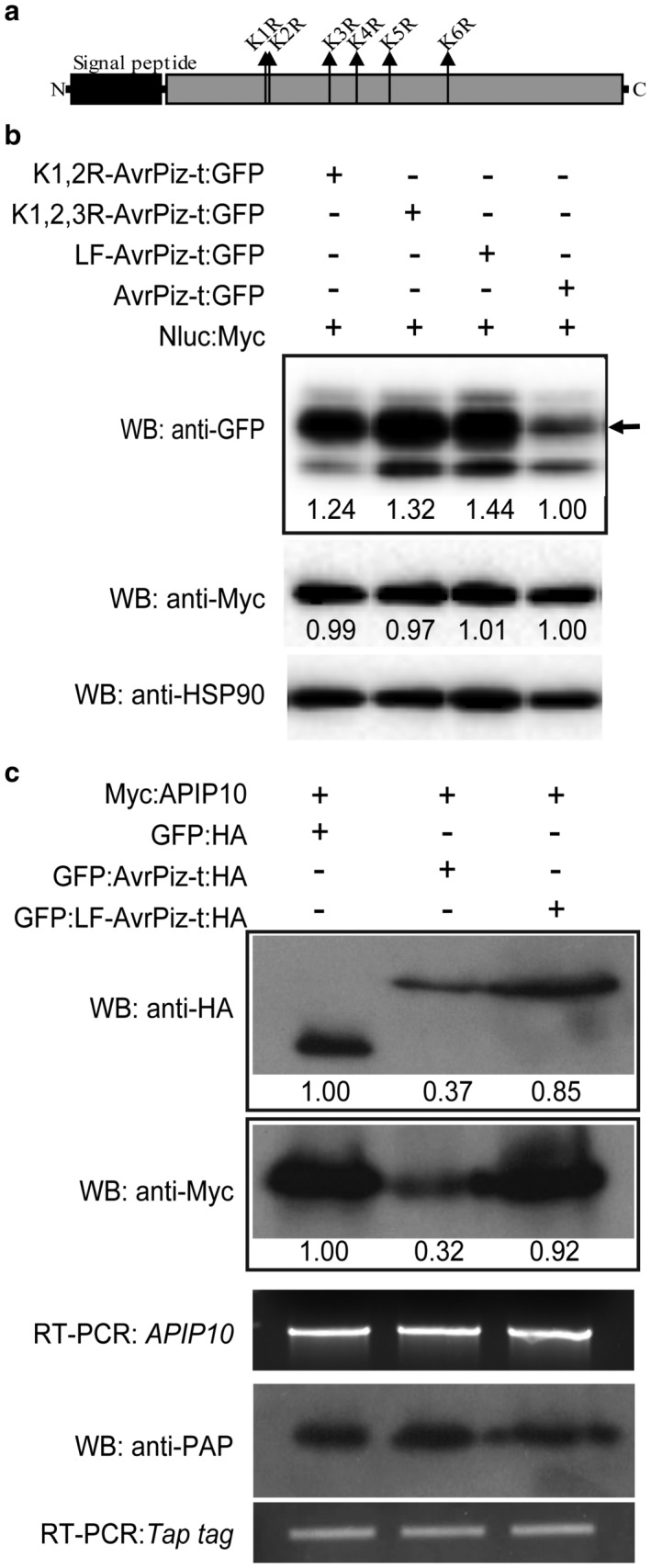
Protein stability of double, triple and lysine‐free mutants of AvrPiz‐t and APIP10 in plant cells. (a) Schematic structure of the AvrPiz‐t protein; the locations of the six lysine residues are labelled as K1–K6 according to their order in the AvrPiz‐t protein. (b) Plasmids of K1,2R‐AvrPiz‐t:GFP, K1,2,3R‐AvrPiz‐t:GFP, LF‐AvrPiz‐t:GFP and AvrPiz‐t:GFP were separately expressed in Nipponbare rice protoplasts, and Nluc:Myc was used as an internal transfection control. Immunoblots with anti‐GFP, anti‐Myc and anti‐HSP90 were conducted to detect the protein levels of different AvrPiz‐t mutants, Nluc‐Myc and endogenous rice HSP90, respectively. (c) Co‐expression of Myc:APIP10 with GFP:LF‐AvrPiz‐t:HA and GFP:AvrPiz‐t:HA in Nicotiana benthamiana. GFP:HA was used as the infiltration control, and the Tap tag was used as an internal control. Semi‐real‐time polymerase chain reaction (semi‐RT‐PCR) was conducted for APIP10 and Tap transcripts. Protein levels were measured by Image Lab software on the basis of the band intensity and were then normalized with the internal control.
