Figure 1.
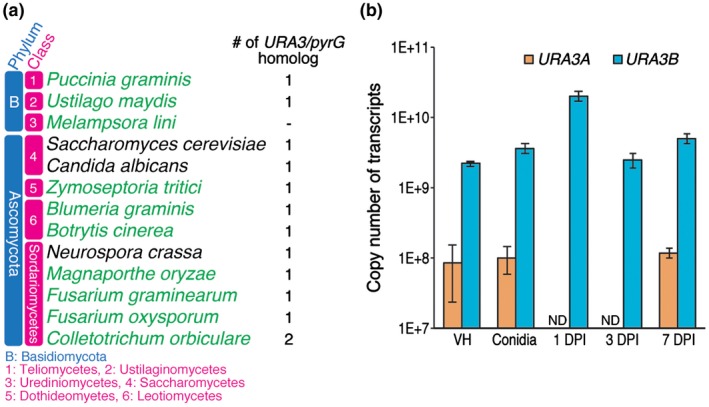
URA3/pyrG homologues are conserved in plant fungal pathogens. (a) The number of predicted URA3/pyrG homologues in selected plant fungal pathogens (green type) and model fungi (black type). Homologues were predicted by BLAST search using the Saccharomyces cerevisiae Ura3p amino acid sequence as a query. (b) URA3A and URA3B expression profiles quantified by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis. Bars represent the absolute number of transcripts. RNAs were extracted from vegetative hyphae (VH), conidia, epidermal cells at 1 day post‐inoculation (DPI), epidermal cells at 3 DPI and whole leaf tissue at 7 DPI. Total RNA levels were normalized using the RIBOSOMAL PROTEIN I5 gene (Cob_11000), as reported previously (Gan et al., 2013). Error bars represent standard errors. n = 3. ND indicates not detected. Primers used to detect URA3A, URA3B and RPI5 are listed in Table S3 (see Supporting Information). [Colour figure can be viewed at wileyonlinelibrary.com]
