Figure 5.
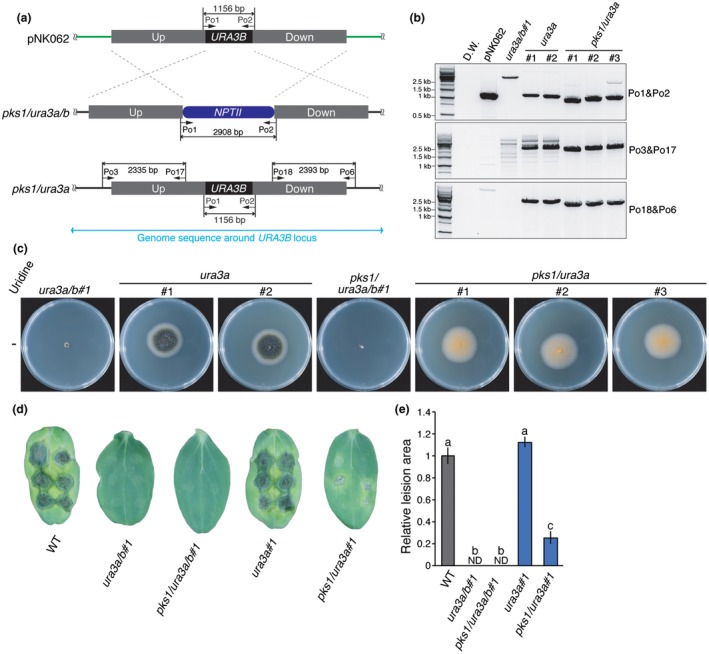
URA3B knock‐in to its original locus enabled in planta virulence assay using the ura3a/b mutant‐based marker recycling system. (a) Schematic diagrams of URA3B knock‐in experiments on the ura3a/b mutant strains. The pNK062 plasmid contains 2 kb of upstream (Up) and downstream (Down) sequences of the URA3B coding sequence (CDS) on the Colletotrichum orbiculare 104‐T genome. The complete URA3B CDS, including intron, is located between the Up and Down sequences. (b) Genomic DNA polymerase chain reaction (PCR) showed that URA3B was knocked in, resulting in ura3a and pks1/ura3a mutants. The primer set Po1/Po2 is described in Fig. 2b. If the URA3B CDS is knocked in to ura3a/b and pks1/ura3a/b mutants, primer sets Po3/Po17 and Po18/Po6 generate 2335‐ and 2393‐bp bands, respectively. The primers used are listed in Table S3 (see Supporting Information). (c) All strains were cultured on potato dextrose agar (PDA) for 6 days at 25 °C in the dark. ura3a#1, ura3a#2, pks1/ura3a#1, pks1/ura3a#2 and pks1/ura3a#3 strains can grow on PDA without uridine addition, demonstrating the knock‐in of URA3B. (d) Conidia of each strain were inoculated onto plants. At 9 days post‐germination, cotyledons of cucumber were inoculated with six drops of 10 µL conidia solution at 5 × 105 conidia/mL. Photographs were taken after 6 days of incubation. The disease symptoms of ura3a#1 were similar to those of the WT strain, showing that URA3B is able to complement the reduced virulence phenotype of the ura3a/b mutant. The reduced virulence phenotype of pks1/ura3a#1 reflects the contribution of PKS1 to the virulence of C. orbiculare. The details of each strain are listed in Table S1 (see Supporting Information). (e) The area of each lesion shown in (d) was measured using Image J software. Values were normalized to set WT as unity. n = 12. Error bars represent standard errors. ND indicates not detected. Different letters on the bars represent significant differences (Tukey’s test, P < 0.01). [Colour figure can be viewed at wileyonlinelibrary.com]
