Summary
Rice black‐streaked dwarf virus (RBSDV), a member of the genus Fijivirus, is a devastating pathogen of crop plants. RBSDV S10 encodes a capsid protein (P10) that is an important component of the double‐layered particle. However, little information is available on the roles of RBSDV P10 in viral infection or in interactions with other viruses. Here, we demonstrate that the expression of P10 in plants alleviates the symptoms of both RBSDV and the closely related Southern rice black‐streaked dwarf virus (SRBSDV), and reduces the disease incidence, but renders the plants more susceptible to the unrelated Rice stripe virus (RSV). Further experiments suggest that P10‐mediated resistance to RBSDV and SRBSDV operates at the protein level, rather than the RNA level, and is not a result of post‐transcriptional gene silencing. Transcriptomic data reveal that the expression of P10 in plants significantly suppresses the expression of rice defence‐related genes, which may play important roles in resistance to RSV infection. After infection with RBSDV, plants are more resistant to subsequent challenge by SRBSDV, but more susceptible to RSV. Overall, these results indicate that P10 acts as an important effector in virus interactions.
Keywords: antagonism, coat protein, coat protein‐mediated resistance, Rice black‐streaked dwarf virus, Rice stripe virus, Southern rice black‐streaked dwarf virus, synergism
Introduction
Rice black‐streaked dwarf virus (RBSDV), belonging to the genus Fijivirus in the family Reoviridae, is transmitted to rice, maize, barley and wheat by the small brown planthopper (Laodelphax striatellus) in a persistent, propagative manner (Wei and Li, 2016). RBSDV was first reported in Japan (Kuribayashi and Shinkai, 1952), and has caused severe economic losses to rice and maize production in Asia. Plants infected by RBSDV display severe growth abnormalities, especially severe stunting (Fang et al., 2000; Shikata and Kitagawa, 1977; Wang et al., 2003). RBSDV is a double‐stranded RNA virus with 10 genome segments (S1–S10). Most segments encode one protein, although S5, S7 and S9 each have two open reading frames (Zhang et al., 2001). Segments S1, S2 and S3 encode an RNA‐dependent RNA polymerase (P1), a core protein (P2) and a capping enzyme (P3), respectively. The P5‐1, P6 and P9‐1 proteins together constitute the viroplasm (Akita et al., 2012; Sun L et al., 2013; Wang et al., 2011), which is the site of viral replication and assembly. P7‐1 is a protein that forms tubules at the plasmodesmata of plant cells (Isogai et al., 1998; Sun Z et al., 2013b). P7‐2 is a component of the SCF complex (Skp, Cullin, F‐box‐containing complex) (Tao et al., 2017). P8 and P10 are a core capsid protein and an outer capsid protein, respectively (Liu et al., 2007a, b; Sun Z et al., 2013a).
Mixed infections of two related or unrelated viral pathogens in the same plant commonly occur under natural conditions (Syller, 2012). In some combinations of viruses, there is an observable influence of one virus on the replication or transmission of the second virus. In a synergistic interaction, there is a positive effect on one or both viruses, resulting in an increase in viral replication or movement in the host plant. Synergistic virus infections usually have more severe effects on crop production than do single infections with either of the individual viruses. Numerous synergistic interactions have been reported (García‐Cano et al., 2006; Li et al., 2014; Untiveros et al., 2007). Synergistic interactions frequently involve unrelated viruses: for example, a mixed infection of Potato virus Y (PVY) and Potato virus X (PVX) results in an enhancement of disease symptoms in tobacco (Bance, 1991; González‐Jara et al., 2004). Mechanisms of synergism are diverse in different host–pathogen systems. For example, PVY HC‐Pro (helper component proteinase) mediates Cucumber mosaic virus (CMV)–PVY synergistic interactions by suppression of post‐transcriptional gene silencing (PTGS) (Fukuzawa et al., 2010). Beet curly top virus (BCTV) C2 protein promotes Tomato yellow leaf curl Sardinia virus (TYLCSV) replication by creating a suitable cell environment (Caracuel et al., 2012).
Antagonistic interactions often occur between related viruses. Antagonism is also referred to as cross‐protection when deployed in agricultural applications, or as superinfection exclusion (Syller and Grupa, 2016; Ziebell and Carr, 2010). This type of interaction occurs when an initial viral infection prevents or interferes with subsequent infection by a similar virus. In the field, cross‐protection involves the pre‐inoculation of crop plants with a mild or attenuated virus strain to prevent subsequent infection by severe strains of the same virus, and has been successfully used to prevent damaging infections of Papaya ringspot virus (PRSV) and Citrus tristeza virus (Folimonova, 2013; Gonsalves, 1998). The mechanistic basis for synergistic interactions between viruses has been explored in a number of studies, but much less is known about the mechanisms involved in antagonistic interactions (Ratcliff et al., 1999; Ziebell and Carr, 2009; Ziebell et al., 2007). Although antiviral RNA silencing may play a vital role in cross‐protection between very closely related strains of the same virus (Ratcliff et al., 1999), there have been some reports that RNA silencing is not solely responsible for the mechanism of cross‐protection. For example, the CMV mutant Fny‐CMVΔ2b, which lacks the 2b silencing suppressor protein, not only provides cross‐protection against wild‐type CMV, but also against the less closely related strain TC‐CMV (Ziebell and Carr, 2009; Ziebell et al., 2007).
Here, we focus on the infection of rice by the widespread, devastating virus RBSDV. Our study looks at co‐infections of rice with RBSDV and either a related reovirus, Southern rice black‐streaked dwarf virus (SRBSDV), or the unrelated Rice stripe virus (Phenuiviridae, Tenuivirus, RSV). SRBSDV is transmitted by the white‐backed planthopper (Sogatella furcifera) in a persistent, circulative–propagative manner (Pu et al., 2012; Zhou G et al., 2013). RSV propagates in, and is transmitted by, the same small brown planthopper (Laodelphax striatellus) which transmits RBSDV. RBSDV, SRBSDV and RSV have all caused serious disease outbreaks recently in southeastern Asia, especially in the rice fields of China (Wei et al., 2009; Zhou G et al., 2013).
In the experiments described here, we demonstrate that plants infected with RBSDV, or those expressing the RBSDV P10 outer capsid protein, are less susceptible to challenge with the related SRBSDV, but more susceptible to the unrelated RSV. These results have implications for both the natural development of mixed infections and the potential use of transgenic plants as a virus control strategy in the field.
Results
Expression of the P10 gene in rice affects plant growth and development
Our previous research has shown that RBSDV P10 is a membrane protein localized to the endoplasmic reticulum (ER) (Sun Z et al., 2013a). Transient expression of P10 induces the expression of ER stress marker genes, indicating that it triggers the ER stress and unfolded protein response. In order to further study the P10 protein, we produced transgenic rice plants expressing the P10 gene (OEP10) driven by the Cauliflower mosaic virus (CaMV) 35S promoter. Two homozygous lines and their T3 generation plants were used in subsequent experiments. The protein expression levels of P10 in OEP10 transgenic plants were verified by western blotting (Fig. 1a). Interestingly, the phenotypes of the plants of the two transgenic lines were similar to that of the control non‐transgenic (NIP) plants before heading, but, at maturity, the transgenic lines were about 20% shorter than the controls (Fig. 1b,c). The transgenic plants produced many more tillers per plant (Fig. 1d) and, at the ripening stage, had a smaller number of grains per panicle (Fig. 1e). These results demonstrate that transgenic expression of the P10 gene in rice affects both the growth and development of the plants.
Figure 1.
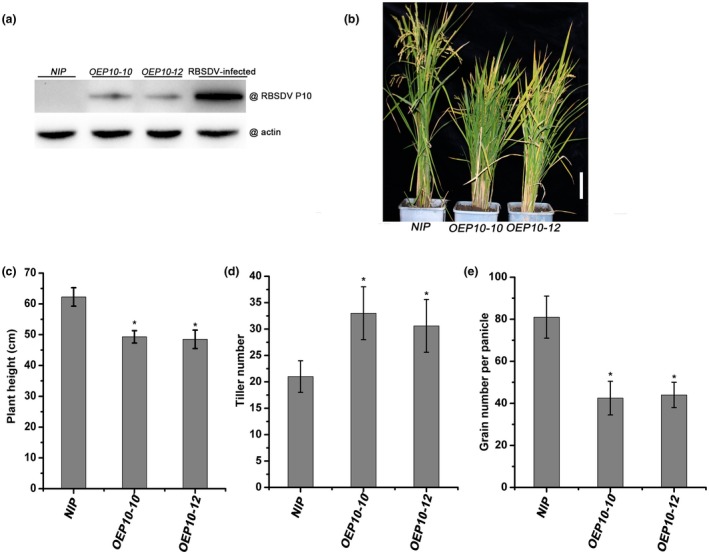
The phenotype of OEP10 transgenic plants expressing the Rice black‐streaked dwarf virus (RBSDV) P10 protein. (a) Western blotting showing P10 protein expression levels in transgenic and RBSDV‐infected plants. (b) Phenotypes of 3‐month‐old NIP (non‐transformed controls), OEP10‐10 and OEP10‐12 plants. White bar represents 10 cm. (c) Heights of NIP, OEP10‐10 and OEP10‐12 plants. (d) Tiller numbers in NIP, OEP10‐10 and OEP10‐12 plants. (e) Grain numbers in each panicle of NIP, OEP10‐10 and OEP10‐12 plants. *Significant difference at P < 0.05.
Expression of the P10 gene in rice plants reduces their susceptibility to RBSDV
To further study the function of P10 in RBSDV infection, we inoculated the OEP10 plants with RBSDV. Both transgenic lines were more resistant than control NIP plants to RBSDV infection. RBSDV‐infected OEP10‐10 and OEP10‐12 plants were less stunted than RBSDV‐infected NIP plants (Figs 2a and S2, see Supporting Information). In addition, we measured the viral RNA levels at different times after RBSDV inoculation. The levels of RBSDV S4, S5, S6, S7 and S10 RNA in both transgenic lines were significantly reduced compared with those in control plants at 10 days post‐inoculation (dpi), and remained at greatly reduced levels for up to 30 dpi (Figs 2b,c and S3, see Supporting Information). As shown in Fig. 2d, the viral incidence in both OEP10‐10 and OEP10‐12 lines (42% and 52%, respectively) was less than that in NIP (83%). Thus, transgenic expression of the P10 gene in rice enhances resistance to RBSDV infection.
Figure 2.
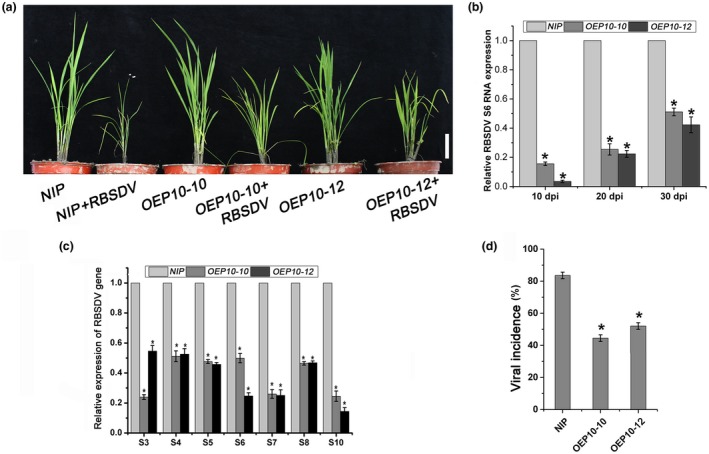
The effects of Rice black‐streaked dwarf virus (RBSDV) infection on OEP10 transgenic plants expressing the RBSDV P10 protein. (a) The visual appearance of mock‐ and RBSDV‐infected NIP (non‐transformed controls), OEP10‐10 and OEP10‐12 plants. White bar represents 5 cm. (b) Quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) measurements of the relative expression levels of RBSDV S6 in RBSDV‐infected OEP10‐10 and OEP10‐12 plants compared with that in RBSDV‐infected NIP at different times. (c) The relative expression levels of RBSDV genomic RNA (S3, S4, S5, S6, S7, S8, S10) levels in RBSDV‐infected OEP10‐10 and OEP10‐12 plants compared with that in RBSDV‐infected NIP as assessed by RT‐qPCR at 30 days post‐inoculation (dpi). Relative transcript levels were analysed using the 2–ΔΔC(t) method. (d) Viral incidence in NIP, OEP10‐10 and OEP10‐12 plants. The numbers of healthy and diseased plants for each treatment were determined by RT‐PCR at 30 dpi. Each treatment used at least 30 seedlings, and three biological replicates were performed. The average values from three biological replicates are shown. Error bars represent ± standard deviation (SD). An asterisk at the top of a column indicates a significant difference at P < 0.05.
Previous reports have indicated that RNA‐mediated interference contributes to coat protein (CP)‐mediated viral resistance (Germundsson et al., 2002; Haan et al., 1992). To examine whether the P10‐mediated RBSDV resistance observed in our transgenic plants was caused by the P10 protein itself, or by the P10 RNA, an additional transgenic rice line (P10 RNA) was generated in which the P10 transgene was altered so that a functional P10 protein could not be expressed. Specifically, the P10 gene was modified to insert a translation termination codon directly following the P10 gene translation initiation codon. The morphology of P10 RNA transgenic plants was similar to that of non‐transgenic plants. The expression level of P10 RNA in transgenic plants was verified by western blotting. The P10 protein could not be detected in P10 RNA plants (Fig. S1, see Supporting Information). T2 generation P10 RNA transgenic plants were inoculated with RBSDV as described above. As shown in Fig. S4 (see Supporting Information), the viral RNA level and viral incidence in P10 RNA transgenic plants were not significantly different from those in non‐transgenic control plants, demonstrating that it is the P10 protein, and not the P10 RNA, that induces partial resistance to RBSDV.
Transgenic expression of the P10 protein could conceivably lead to a reduction in RBSDV infection by interfering with the small RNA‐directed anti‐virus defence response. To examine this possibility, we performed high‐throughput sequencing of small RNAs from transgenic and non‐transgenic plants inoculated with the virus. As shown in Table S2 (see Supporting Information), the size distributions of virus‐derived small interfering RNAs (vsiRNAs) in RBSDV‐infected NIP were mostly 21 and 22 nucleotides long, similar to previous reports (Lan et al., 2017; Sun et al., 2015). vsiRNAs were rarely detected in either transgenic or non‐transgenic plants (Table S2). More importantly, there were approximately 60% fewer vsiRNAs in OEP10‐10 plants than in non‐transgenic NIP plants (Fig. 3 and Table S2). These results show that both the S10 RNA level (Fig. 2c) and its siRNA abundance in RBSDV‐infected OEP10‐10 plants are lower than those in RBSDV‐infected NIP plants. As RNA levels and siRNA abundance are negatively correlated, we conclude that the vsiRNA‐mediated antiviral mechanism is unlikely to be responsible for the P10‐induced partial resistance to RBSDV.
Figure 3.
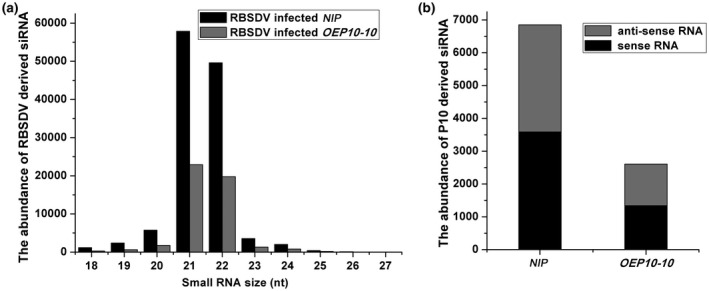
The abundance of virus‐derived small interfering RNAs (vsiRNAs) in Rice black‐streaked dwarf virus (RBSDV)‐infected plants. (a) The size distribution (nucleotides, nt) of RBSDV‐derived siRNAs in RBSDV‐infected plants. (b) The abundance of P10‐derived siRNAs in RBSDV‐infected plants.
Expression of the P10 gene in rice plants reduces their susceptibility to SRBSDV
Because SRBSDV is a fijivirus closely related to RBSDV, we next investigated whether OEP10 plants exhibited resistance to SRBSDV infection in the same way as to RBSDV. The transgenic OEP10‐10 and control NIP plants were infested with virus‐free and SRBSDV‐infected white‐backed planthoppers using seven insects per seedling. All planthoppers were removed after a 3‐day feeding period. As shown in Fig. 4a, SRBSDV‐infected NIP plants were more dwarfed and displayed more severe symptoms than did SRBSDV‐infected transgenic OEP10‐10 plants. Real‐time RT‐PCR showed that SRBSDV RNA levels in OEP10‐10 plants were less than 20% of those in non‐transgenic controls (Fig. 4b). A similar difference was noted when P8 protein levels were examined by western blotting (Fig. 4c). In addition, the incidence of viral infection was significantly higher in NIP plants (78%) relative to OEP10‐10 plants (60%) (Fig. 4d). As shown in Fig. S5 (see Supporting Information), the viral RNA levels and viral incidence in OEP10‐12 transgenic plants were similar to those in OEP10‐10 plants in response to SRBSDV infection. These results confirm that transgenic OEP10 plants expressing the RBSDV P10 protein show enhanced resistance to SRBSDV infection relative to NIP plants.
Figure 4.
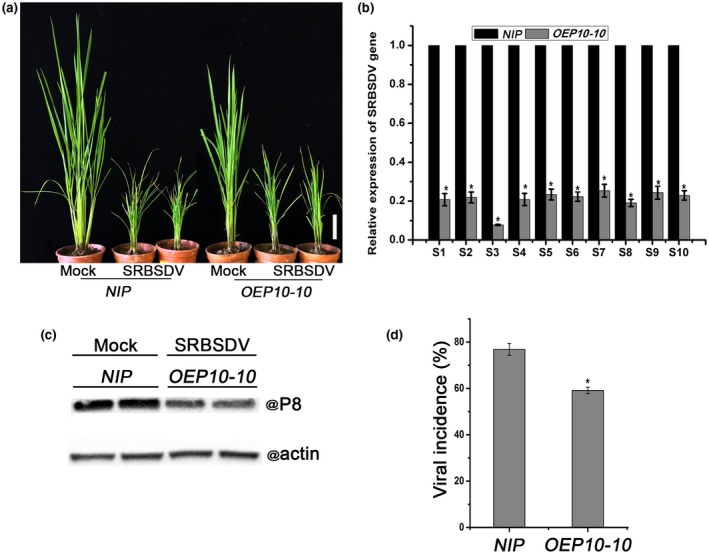
The effect of Southern rice black‐streaked dwarf virus (SRBSDV) infection on OEP10 transgenic plants expressing the Rice black‐streaked dwarf virus (RBSDV) P10 protein. (a) The visual appearance of mock‐ and SRBSDV‐infected plants. White bar represents 5 cm. (b) The relative expression levels of SRBSDV genes in SRBSDV‐infected plants as assessed by quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR). (c) SRBSDV protein levels in SRBSDV‐infected plants determined by western blotting. (d) Viral incidence in NIP (non‐transformed controls) and OEP10‐10 plants. The numbers of healthy and diseased plants in each treatment were determined by RT‐PCR at 30 days post‐inoculation. Each treatment used at least 30 seedlings, and three biological replicates were performed. Error bars represent ± standard deviation (SD). An asterisk at the top of a column indicates significant difference at P < 0.05.
There is mutual antagonism between RBSDV and SRBSDV in sequential infections
Because P10 enhanced the resistance of rice to both RBSDV and SRBSDV, we next tested whether there is antagonism between RBSDV and SRBSDV in sequential infections. Rice seedlings were first inoculated with RBSDV using viruliferous small brown planthoppers. Virus‐free planthoppers were used for the controls. After 7–10 days, the seedlings were inoculated with SRBSDV. Plants singly infected with RBSDV or SRBSDV were used as controls. Each treatment had two replicates each of 20–30 seedlings, and plants were then tested for viruses by RT‐PCR using primers specific for RBSDV and SRBSDV, as described previously (Cheng et al., 2013). As shown in Table 1, most plants inoculated using planthoppers carrying either RBSDV or SRBSDV became infected with the respective virus. However, when insects carrying SRBSDV were fed on plants that had first been inoculated with RBSDV, most plants became infected only with RBSDV, none became infected with both RBSDV and SRBSDV, and the few plants that did develop SRBSDV infection were free from RBSDV. Similarly, in a reciprocal experiment in which plants were first inoculated with SRBSDV and then challenged with RBSDV, no plants were obtained that had a mixed infection of both viruses (Table 2). These results therefore indicate a mutual antagonism between RBSDV and SRBSDV irrespective of the order of infection.
Table 1.
The effect of Rice black‐streaked dwarf virus (RBSDV) on subsequent infection by Southern rice black‐streaked dwarf virus (SRBSDV).
| Experiment I | Experiment II | |||||||
|---|---|---|---|---|---|---|---|---|
| Total* | RB† | SRB‡ | RB‐SRB§ | Total | RB | SRB | RB‐SRB | |
| RB single inoculation | 20* | 20 | – | – | 18 | 15 | – | – |
| SRB single inoculation | 18 | – | 18 | – | 16 | – | 16 | – |
| Primary inoculation by RB and secondary inoculation by SRB | 24 | 24 | 0 | 0 | 22 | 18 | 4 | 0 |
Total is the number of inoculated seedlings.
RB is the number of RBSDV‐infected plants.
SRB is the number of SRBSDV‐infected plants.
RB‐SRB is the number of plants co‐infected with both RBSDV and SRBSDV.
Table 2.
The effect of Southern rice black‐streaked dwarf virus (SRBSDV) on subsequent infection by Rice black‐streaked dwarf virus (RBSDV).
| Experiment I | Experiment II | |||||||
|---|---|---|---|---|---|---|---|---|
| Total* | SRB† | RB‡ | SRB‐RB§ | Total | SRB | RB | SRB‐RB | |
| SRB single inoculation | 16 | 10 | ‐ | ‐ | 20 | 15 | ‐ | ‐ |
| RB single inoculation | 18 | ‐ | 9 | ‐ | 20 | ‐ | 11 | ‐ |
| primary inoculation by SRB and secondary inoculation by RB | 18 | 12 | 1 | 0 | 25 | 18 | 2 | 0 |
Total is the number of inoculated seedlings.
SRB is the number of SRBSDV‐infected plants.
RB is the number of RBSDV‐infected plants.
SRB‐RB is the number of plants co‐infected with both SRBSDV and RBSDV.
Expression of the P10 gene in transgenic rice plants, or prior infection by RBSDV, increases susceptibility to RSV
To investigate how RBSDV P10 expression would affect infection by an unrelated virus, we challenged the two transgenic lines with the tenuivirus RSV using viruliferous small brown planthoppers. As shown in Fig. 5a, the symptoms in RSV‐infected control NIP plants were necrotic stripes, plant wilting and stunting. However, RSV‐infected OEP10‐10 plants showed more necrosis, increased wilting and more severe stunting. The viral incidence in OEP10‐10 plants (63%) was significantly greater than that in non‐transgenic NIP plants (41%) (Fig. 5b). Real‐time RT‐PCR measurement showed that the levels of RSV CP RNA in RSV‐infected OEP10‐10 plants were four‐ to five‐fold higher than those in RSV‐infected NIP plants (Fig. 5c). Similar results were observed for the accumulation of the RSV CP using western blotting (Fig. 5d). Similar results were obtained using transgenic OEP10‐12 plants (Fig. S6, see Supporting Information). These results suggest that the expression of RBSDV P10 in rice plants renders them more susceptible to RSV.
Figure 5.
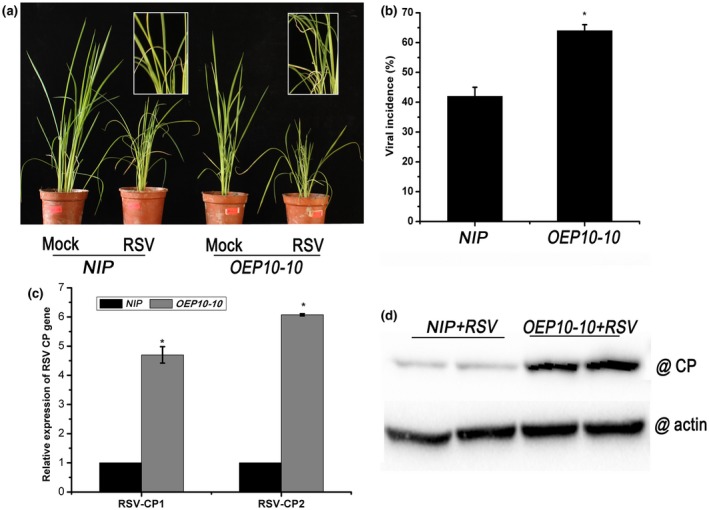
The effect of Rice stripe virus (RSV) infection on OEP10 transgenic plants expressing the Rice black‐streaked dwarf virus (RBSDV) P10 protein. (a) The visual appearance of mock‐ and RSV‐infected plants. (b) Viral incidence in NIP (non‐transformed controls) and OEP10‐10 plants. The numbers of healthy and diseased plants in each treatment were determined by reverse transcription‐polymerase chain reaction (RT‐PCR) at 30 days post‐inoculation. Each treatment used at least 30 seedlings, and three biological replicates were performed. (c) The relative expression levels of the RSV coat protein (CP) gene in RSV‐infected plants assessed by quantitative RT‐PCR. RSV‐CP1 and RSV‐CP2 are two pairs of primers designed at different positions in the RSV CP gene. (d) The RSV CP protein levels in RSV‐infected plants determined by western blotting. Error bars represent ± standard deviation (SD). An asterisk at the top of a column indicates a significant difference at P < 0.05.
In natural conditions, both RBSDV and RSV are transmitted by the same vector, and natural co‐infection of rice by both viruses is common (Cho et al., 2013; Li et al., 2015). As P10 expression increased susceptibility to RSV, we next examined whether RBSDV itself interacts with RSV in a synergistic relationship. Rice seedlings were first inoculated with RBSDV using viruliferous small brown planthoppers and, 7 days later, these were removed and replaced with planthoppers transmitting RSV. Virus symptoms were observed at about 20 days after RSV inoculation. As shown in Fig. 6a, infection by either RBSDV or RSV alone in plants resulted in dwarfism and the development of necrotic stripes in leaves. However, more severe symptoms were observed in plants inoculated with both viruses. The relative levels of RBSDV and RSV genomic RNAs and CPs were monitored by real‐time RT‐PCR and western blotting, respectively. Consistent with the symptoms, the RBSDV and RSV RNA levels in dual‐infected rice were two‐ to three‐fold higher than those in singly infected plants (Fig. 6b,c). Similar results were observed with regard to viral protein expression levels (Fig. 6d,e). These results confirm that a synergistic interaction occurs between RBSDV and RSV, with the accumulation of both viruses being increased in the dual infection.
Figure 6.
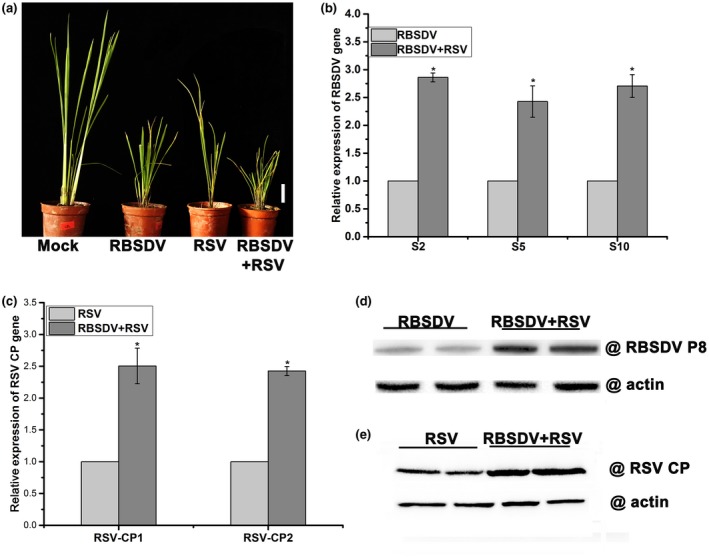
The synergistic interaction between Rice black‐streaked dwarf virus (RBSDV) and Rice stripe virus (RSV). (a) The symptoms in mock‐, RBSDV‐, RSV‐ and jointly infected plants. White bar represents 5 cm. (b) The relative expression levels of RBSDV genomic RNAs (S2, S5 and S10) in plants infected with RBSDV only or jointly with RBSDV and RSV. (c) The relative expression levels of RSV genes in plants infected with RSV only or jointly with RBSDV and RSV as shown by quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR); RSV‐CP1 and RSV‐CP2 are two pairs of primers designed at different positions in the RSV coat protein (CP) gene. (d) RBSDV P8 protein levels in plants infected with RBSDV only or with RBSDV and RSV as assessed by western blotting. (e) The RSV CP protein levels in plants infected with RSV only or with both RBSDV and RSV as assessed by western blotting. Error bars represent ± standard deviation (SD). An asterisk at the top of a column indicates a significant difference at P < 0.05.
When RSV was inoculated first and the plants were then challenged by RBSDV, there was a different pattern of response. In the jointly infected plants, RSV titres were significantly greater than in plants infected with only RSV, but RBSDV titres were markedly lower than in those infected with only RBSDV (Fig. S8, see Supporting Information). Thus, the dual infection is always in favour of RSV, whereas the infection order determines the outcome of RBSDV multiplication.
Transcriptome analysis of rice expressing the RBSDV P10 protein
To shed light on the effect of expression of the RBSDV P10 protein in rice, we compared the global transcriptome of the transgenic OEP10‐10 line with that of non‐transgenic rice plants (NIP). Ten‐day‐old rice seedlings were collected for RNA extraction and RNA sequencing. A total of 756 genes (523 repressed and 233 induced) were differentially expressed (two‐fold change, P ≤ 0.05) (Fig. S7a, see Supporting Information). A list of the genes identified is presented in Table S3 (see Supporting Information). KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis confirmed that P10 mainly affected plant–pathogen interaction, plant hormone signal transduction, ascorbate and aldarate metabolism and secondary metabolism, including stilbenoid biosynthesis, phenylpropanoid biosynthesis, limonene and pinene degradation and flavonoid biosynthesis pathways (Fig. S7b). For the plant–pathogen interaction pathway, the expression of 49 genes was significantly changed in the OEP10‐10 line. The accuracy of the transcriptomic data for these defence genes was verified by quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR). As shown in Fig. 7, the plant defence‐related genes, such as WRKY transcription factors and JAZ genes, were suppressed. In addition, the receptor kinase and OsWAK family belonging to the receptor signalling pathway were also altered significantly (Table 3). These results suggest that the expression of P10 in rice affects plant defence responses. In the plant hormone signalling pathways, the genes involved in auxin, cytokinin (CK) and brassinosteroid (BR) pathways were changed significantly (Table 3). These genes are involved in plant growth and development, and thus these changes are consistent with the dwarfism and developmental abnormalities seen in the P10 transgenic plants.
Figure 7.
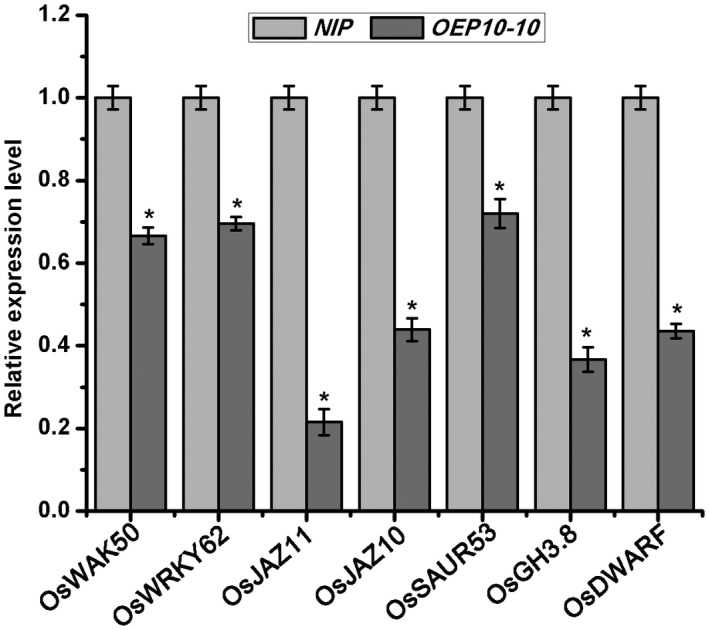
Quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) data showing the expression levels of defence response genes in OEP10 transgenic plants expressing the Rice black‐streaked dwarf virus (RBSDV) P10 protein relative to those in the non‐transgenic NIP controls. UBQ5 was used as the internal reference gene. Data are means ± standard deviation (SD) from three biological replicates. An asterisk indicates significant difference between NIP and OEP10 transgenic plants at P ≤ 0.05.
Table 3.
Response of plant hormone pathway genes to expression of P10 in rice.
| Gene ID | Gene description | Log2(FC)* |
|---|---|---|
| Plant–pathogen interaction | ||
| LOC_Os01g60640 | WRKY21 | −2.30123 |
| LOC_Os09g25070 | WRKY62 | −2.02107 |
| LOC_Os11g02520 | WRKY104 | −1.70669 |
| LOC_Os05g25770 | WRKY45 | −1.63021 |
| LOC_Os05g39720 | WRKY70 | −1.33889 |
| LOC_Os03g53050 | WRKY121 | −1.32924 |
| LOC_Os11g02480 | WRKY46 | −1.26024 |
| LOC_Os09g16510 | WRKY74 | −1.24785 |
| LOC_Os01g61080 | WRKY24 | −1.22253 |
| LOC_Os03g28940 | JAZ6 | −1.10866 |
| LOC_Os03g08320 | JAZ11 | −1.85962 |
| LOC_Os10g25290 | JAZ12 | −1.71394 |
| LOC_Os03g08330 | JAZ10 | −2.04555 |
| Toll‐like receptor signalling pathway | ||
| LOC_Os04g51040 | OsWAK50 | −2.09454 |
| LOC_Os04g29580 | OsWAK37 | −1.54263 |
| LOC_Os02g56370 | OsWAK20 | −1.47422 |
| LOC_Os02g02120 | OsWAK11 | −1.43851 |
| LOC_Os04g29960 | OsWAK43 | −1.40899 |
| LOC_Os01g26280 | OsWAK8 | 1.109275 |
| LOC_Os04g30240 | OsWAK60 | 1.235246 |
| LOC_Os04g29770 | OsWAK3 | 1.32052 |
| LOC_Os04g21790 | OsWAK34 | 1.544207 |
| LOC_Os10g10130 | OsWAK112d | 1.54711 |
| LOC_Os12g42070 | OsWAK129b | 1.722076 |
| LOC_Os04g29680 | OsWAK38 | 2.276714 |
| LOC_Os09g38840 | OsWAK90 | 5.244784 |
| Auxin pathway | ||
| LOC_Os09g37480 | OsSAUR53 | −3.24823 |
| LOC_Os02g52990 | OsSAUR12 | −3.03287 |
| LOC_Os07g40290 | OsGH3.8 | −2.20391 |
| LOC_Os06g48950 | Auxin response factor 19 | −1.46253 |
| LOC_Os01g12160 | OsGH3.3 | 1.442901 |
| LOC_Os09g37330 | OsSAUR39 | 1.576482 |
| LOC_Os01g45550 | Auxin efflux carrier component | −2.07308 |
| LOC_Os11g44810 | Auxin‐repressed protein | 1.880417 |
| LOC_Os05g41420 | Auxin‐induced protein 5NG4 | 3.915879 |
| Cytokinin pathway | ||
| LOC_Os01g56810 | Cytokinin dehydrogenase | −4.2629 |
| LOC_Os08g35860 | Cytokinin dehydrogenase | 1.132087 |
| LOC_Os05g42040 | UDP‐glucoronosyl and UDP‐glucosyl transferase | −1.43682 |
| LOC_Os05g42060 | UDP‐glucoronosyl and UDP‐glucosyl transferase | −1.27073 |
| LOC_Os10g09990 | Cytokinin‐O‐glucosyltransferase | 2.581396 |
| LOC_Os11g04720 | OsRR10 type‐A response | 2.618383 |
| Brassinosteroid pathway | ||
| LOC_Os03g40540 | OsDWARF | −2.32283 |
| LOC_Os07g44130 | Cytochrome P450 72A1 | −1.02609 |
| LOC_Os06g02019 | Cytochrome P450 | 1.628058 |
| LOC_Os07g33480 | Cytochrome P450 | 1.74535 |
FC, fold change.
Transcriptomic data obtained from the comparison of OEP10‐10 transgenic plants expressing the Rice black‐streaked dwarf virus (RBSDV) P10 protein with NIP (non‐transformed controls).
Discussion
RBSDV infection causes plant dwarfism and growth abnormalities, symptoms that are often associated with a change in hormone homoeostasis (He et al., 2017). In a previous report, we showed that RBSDV infection affected plant hormones, including the jasmonic acid (JA) and BR pathways. In the present study, we found that expression of the RBSDV P10 protein in plants mimics these morphological changes. Transcriptome analysis indicated that expression of the P10 protein can alter the expression of genes involved in many plant biochemical pathways, including, for example, auxin, CK and BR pathways. This may be the mechanism by which infection of plants by RBSDV induces its characteristic symptoms.
In nature, the co‐infection of rice by RBSDV and SRBSDV has not been reported (Cheng et al., 2013). In this study, it was not possible to superinfect RBSDV‐infected plants with SRBSDV. This phenomenon has been referred to previously as superinfection exclusion or cross‐protection (Syller and Grupa, 2016), and occurs between different strains of the same virus or sometimes between closely related viruses. Although the superinfection exclusion phenomenon has been observed with many plant‐infecting viruses (Capote et al., 2006; Folimonova et al., 2010; Tatineni and French, 2016; Valkonen et al., 2002), only a few viral determinants involved in superinfection exclusion have been identified (Tatineni and French, 2016; Zhang et al., 2017). In some studies, RNA silencing as a result of nucleotide sequence homology between pairs of viruses appears to be the mechanism underlying superinfection exclusion (Valkonen et al., 2002). However, for Wheat streak mosaic virus (WSMV), the NIa‐Pro and CP proteins, but not their RNAs, were the determinants of superinfection exclusion (Tatineni and French, 2016). An alternative model for superinfection exclusion among plant viruses is coat protein‐mediated resistance (CPMR), a term referring to the resistance of transgenic plants expressing virus CP against infection by the same virus or similar virus strains (Beachy et al., 1990; Lindbo and Falk, 2017). CPMR was first demonstrated by the resistance to Tobacco mosaic virus (TMV) of tobacco plants engineered to express the TMV CP gene (Abel et al., 1986). Despite extensive studies, the molecular mechanism of CPMR is not fully understood, but it appears to differ depending on the virus. In the case of CPMR against TMV, the CP is known to interfere with virus particle disassembly, thereby preventing viral RNA replication (Asurmendi et al., 2007; Bendahmane et al., 2007). Interestingly, OEP10 transgenic plants showed some partial resistance to SRBSDV (Fig. 4). It is possible that the P10 protein is not the sole determinant of the RBSDV–SRBSDV antagonism, or that P10 expression levels in transgenic plants were lower than those in RBSDV‐infected plants.
In our study, the expression of a wild‐type P10 gene in rice plants reduced their susceptibility to RBSDV and to the related virus SRBSDV. In contrast, transgenic plants expressing a mutated P10 gene that could not be translated did not exhibit RBSDV resistance. These results suggest a P10 protein‐mediated resistance mechanism, rather than one based on nucleotide sequence homology triggering RNA silencing. Small RNA high‐throughput sequencing results suggested that siRNAs derived from the RBSDV P10 gene were very rarely detected in OEP10‐10 plants. The RNA levels and siRNA abundance of P10 were both significantly reduced in RBSDV‐infected OEP10‐10 plants compared with RBSDV‐infected NIP controls (Fig. 3 and Table S2). These results show that the OEP10‐10 plants were partially resistant to RBSDV, but that this was not a result of the production of more P10‐derived or vsiRNA.
There was a synergistic interaction between the two unrelated viruses RBSDV and RSV: jointly infected plants showed an increased accumulation of RSV and of both viruses when RBSDV was inoculated first. RSV can be transmitted from female hopper adults to their progeny via eggs, whereas RBSDV does not show this transovarial transmission (Wei and Li, 2016). These two viruses cause severe yield losses and significant economic damage, and have been found in mixed infections in commercial crops (Li et al., 2015). Both viruses infect and are transmitted by the same planthopper vector and, in glasshouse experiments, insects co‐infected with both viruses have been found (Li et al., 2013). The synergism detected in the RBSDV–RSV superinfection, leading to an increase in both viruses in the plant, may increase the frequency/efficiency with which the insect vector can acquire each virus. This will probably increase the frequency with which the viruses are transmitted to new, uninfected plants, and so help to spread the infection of both viruses within the crop. However, a different scenario was found when RSV was infected first and RBSDV was inoculated later, suggesting that the infection order determines the outcome of the RBSDV–RSV interaction. A similar effect has been reported for PRSV and Papaya mosaic virus (PapMV). Synergism occurred in plants inoculated first with PRSV and later with PapMV, but there was antagonism if PapMV was inoculated first because the plant defence response was activated against PRSV (Chávez‐Calvillo et al., 2016). We have not studied this, but it is possible that RSV or RSV proteins activate the plant defence response against subsequent RBSDV infection.
Viral synergistic interactions are common in mixed infections of plant viruses, and potential mechanisms behind some of these have been described (Latham and Wilson, 2008). The best‐characterized example is the interaction between PVY and PVX in tobacco plants, resulting in more severe disease symptoms (Bance, 1991). In rice, a synergistic interaction was demonstrated between SRBSDV and Rice ragged stunt virus (Li et al., 2014): co‐infected plants showed aggravated symptoms and increased virus titres. The mechanisms of such synergism are diverse, depending on the host–pathogen systems (Latham and Wilson, 2008). For example, the RNA silencing suppressor HC‐Pro encoded by PVY mediates PVY–CMV synergistic interactions by suppression of PTGS (Fukuzawa et al., 2010). The movement proteins of Red clover mottle virus and TMV can complement one another and promote synergism between these viruses. The White clover yellow vein virus (potyvirus) movement protein P3N‐PIPO facilitates the systemic spread of the White clover mosaic virus (potexvirus) without suppressing RNA silencing (Hisa et al., 2014). The BCTV C2 protein is involved in the promotion of TYLCSV replication by creating a beneficial cell environment for viral spread (Akita et al., 2012). In the present study, we found that transgenic plants expressing the RBSDV P10 protein exhibited more severe symptoms on RSV infection relative to RSV‐infected non‐transgenic control plants (Fig. 5). Transcriptome analysis and RT‐qPCR data revealed that the P10 protein affected the expression of various plant defence pathway components. KEGG pathways belonging to plant–pathogen interactions and plant hormone signal transduction networks were significantly altered. The WRKY transcription factors and JAZ proteins, which are important regulators of salicylic acid (SA) and JA signalling pathways, were significantly suppressed. Previous research has shown that SA‐mediated defence systems are involved in the suppression of RSV multiplication (Wang et al., 2014). We have shown here that P10 inhibition of the plant defence response is important for both RBSDV and RSV infection and replication or movement. These results suggest that P10 is an important effector of the RBSDV–RSV synergistic interaction.
One important outcome of our work is the realization that, although transgenic approaches to virus control may have beneficial effects, as shown by the reduction in susceptibility of P10 plants to RBSDV and SRBSDV, there may also be unintended side‐effects, such as the increase in susceptibility of these plants to RSV.
Experimental Procedures
Plant materials and vectors
Rice cultivars Oryza sativa L. japonica. Huaidao No. 5 and Wuyujing No. 3 were used in this study. Huaidao No. 5 is highly susceptible to RBSDV, SRBSDV and RSV. Cultivar Nipponbare, which is susceptible to these viruses, was used to produce transgenic rice. RBSDV was initially collected from the field and then sequenced by our laboratory [National Center for Biotechnology Information (NCBI) reference, PRJNA14790]. RSV‐infected plants were kindly provided by Professor Yijun Zhou (Jiangsu Academy of Agricultural Sciences, China). Isolates of SRBSDV were kindly provided by Professor Guohui Zhou (South China Agricultural University, China). RBSDV and RSV were transmitted experimentally to rice plants by the small brown planthopper (Laodelphax striatellus), whereas SRBSDV was transmitted by the white‐backed planthopper (Sogatella furcifera). A small brown planthopper population was provided by the Jiangsu Academy of Agricultural Science. Viruliferous or virus‐free planthoppers were reared on healthy rice seedlings (Wuyujing No. 3) in glass beakers at 25 °C. All plants were grown in a glasshouse at 28–30 °C with a 14‐h light/10‐h dark cycle under artificial light.
Construction of overexpression vector and rice transformation
To construct the transgenic expression vector for RBSDV P10, the RBSDV P10 gene or the mutant of P10 (P10RNA), carrying a translation stop codon immediately downstream of the initiation (AUG) codon, was amplified using forward and reverse primers containing BamHI and SacI sites, respectively, as listed in Table S1 (see Supporting Information). The pCV1300 vector which contained the doubled CaMV 35S promoter to drive transcription was digested with BamHI and SacI (Sun Z et al., 2013a). The PCR product was inserted into the vector, and the selected clone was identified by sequencing. The recombinant binary plasmids were introduced into Agrobacterium tumefaciens (strain EHA105) using electroporation and transformed into rice cv. Nipponbare as described previously (Zhou J et al., 2013).
Virus inoculation assay
Inoculation of plants with RBSDV and RSV using small brown planthoppers was performed as described previously, with some modifications (He et al., 2017; Zhou et al., 2010). Three insect nymphs (either viruliferous or virus‐free) were allowed to feed on each plant for 3 days, after which the planthoppers were removed. Then, the plants were further grown in the glasshouse for symptom development to occur. The incidence of small brown planthopper infection with RBSDV or RSV was detected by a dot immunobinding assay (DIBA) (Wu et al., 2013). To acquire SRBSDV, fourth‐stage nymphs of the white‐backed planthopper were fed on SRBSDV‐infected rice plants for 3–4 days. Then, three nymphs were transferred onto Wuyujing seedlings for 10 days to allow the virus to circulate through the insect. For virus‐free nymphs, insects were subjected to a similar process, but were fed on uninfected plants. Rice seedlings at the two‐ to three‐leaf stage were inoculated with three viruliferous or virus‐free nymphs per plant for 3 days and then the planthoppers were removed (Zhou G et al., 2013). The infection of plants with RBSDV, SRBSDV or RSV was confirmed at 30 dpi by RT‐PCR and western blotting. Virus‐specific primers for the detection of RBSDV, SRBSDV and RSV have been described previously (Cheng et al., 2013) and are listed in Table S1. The percentage of plants infected by virus (viral incidence) was determined following RT‐PCR of samples of each plant using virus‐specific primers (Table S1).
RNA extraction and real‐time PCR
Total RNA was extracted from leaves of 30‐day‐old seedlings using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Reverse transcription was performed using the Tiangen fast quant RT kit (Tiangen Company, Beijing, China) with 1–2 μg of total RNA in a 10‐μL reaction. RT‐qPCR assays were performed with ChamQ™ SYBR qPCR Master Mix (Low ROX Premixed), as recommended by the manufacturer, using an ABI7900HT Sequence Detection System (Applied Biosystems, Carlsbad, CA, USA). The RT‐qPCR conditions were as follows: 95 °C for 4 min; 40 cycles of 95 °C for 10 s and 60 °C for 30 s. Relative transcript levels were analysed using the 2–ΔΔC(t) method (Livak and Schmittgen, 2001) by selecting rice UBQ5 as an internal control (Sun et al., 2015). Each experimental result was derived from not less than three biological repeats. Each biological sample consisted of 12–15 pooled plants and was evaluated with five technical replicates. The RT‐qPCR primer sequences used in this study are listed in Table S1.
Western blot analysis
Total protein was extracted from rice leaves with sodium dodecylsulfate (SDS) lysis buffer (100 mm Tris‐HCl, pH 6.8, 10% SDS). Then, 1 μL of 5 × SDS‐polyacrylamide gel electrophoresis (PAGE) sample loading buffer (1 m Tris‐HCl, pH 6.8, 10% SDS, 1% bromophenol blue, 50% glycerine, 2% β‐mercaptoethanol) was added to 4 μL of protein sample, and boiled at 95 °C for 10 min. The protein was separated on 10%–12% SDS‐PAGE gel and transferred to a poly(vinylidene difluoride) (PVDF) membrane. The membrane was blocked with 10% skimmed milk powder diluted in TBST (Tris‐HCl 10mm, pH = 7.4; NaCl 150mm; Tween‐20 0.05%) for 1–2 h. The primary antibody was diluted (1 : 5000) in blocking buffer (5% skimmed milk powder diluted in TBST) for 2 h. Then, secondary antibody was added in blocking buffer (1 : 10 000) and incubated according to the manufacturer’s instructions for 2 h. Antibody binding was detected using a chemiluminescent substrate (ECL; Pierce, Rockford, IL, USA). Anti‐P8 polyclonal antibody (Xie et al., 2017) was used for the diagnosis of RBSDV‐infected seedlings. Anti‐RSV‐CP antibody (provided by Professor Jianxiang Wu) was used for the diagnosis of RSV‐infected rice seedlings. The actin antibody was used for the diagnosis of the reference protein (Abbkine, A01050‐3).
Small RNA and RNA library construction and sequencing
The methods of small RNA and total RNA library construction have been described previously (He et al., 2017; Sun et al., 2015). The rice leaf samples were collected and ground immediately in liquid nitrogen. Total RNAs were extracted from leaves using the TRIzol protocol (Invitrogen). For RNA sequencing, the quantity and purity of total RNA, addition of adapters, size selection and RNA sequencing were performed by Zhejiang Tianke (Hangzhou, China). The total RNA sequencing library was sequenced on an Illumina HiSeq™ 2000 platform (Zhejiang Tianke Company, Hangzhou, China). The mapping of sequencing reads onto the rice genome (The MSU Rice Genome Annotation Project Database Version 7.0) was conducted using Bowtie software. The gene ontology (GO) functional classes and pathways for each sequence were determined using the Blast2go program. A difference in gene expression was considered to be significant when the absolute value of the log2(fold change) ratio was ≥1 and P ≤ 0.05.
Statistical analysis
Differences were analysed using a one‐way analysis of variance (ANOVA) with Fisher’s least significant difference tests. P ≤ 0.05 was considered to be statistically significant. All analyses were performed using ORIGIN 8 software.
Conflicts of Interest
The authors declare no competing financial interests.
Supporting information
Fig. S1 Western blotting to detect P10 protein expression in OEP10 and P10RNA transgenic plants.
Fig. S2 The height of mock‐ and Rice black‐streaked dwarf virus (RBSDV)‐infected (+RB) NIP, OEP10‐10 and OEP10‐12 plants. Error bars indicate ± standard deviation (SD).
Fig. S3 The relative expression levels of some Rice black‐streaked dwarf virus (RBSDV) RNAs in RBSDV‐infected NIP, OEP10‐10 and OEP10‐12 plants. RNA levels of S4, S5, S7 and S10 were tested by quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) at different times in RBSDV‐infected plants. Error bars indicate ± standard deviation (SD). An asterisk at the top of a column indicates a significant difference at P < 0.05.
Fig. S4 (A) Quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) results showing the expression levels of Rice black‐streaked dwarf virus (RBSDV) RNA segments S5, S6 and S8 in RBSDV‐infected P10 RNA transgenic plants relative to the non‐transformed NIP controls at 30 days post‐inoculation (dpi). (B) RBSDV incidence (% plants infected) in NIPand P10 RNA plants. Error bars indicate ± standard deviation (SD).
Fig. S5 (A) Quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) results showing the expression levels of Southern rice black‐streaked dwarf virus (SRBSDV) segments S1–S10 in SRBSDV‐infected OEP10‐12 transgenic plants relative to the non‐transformed NIP controls at 30 days post‐inoculation (dpi). (B) SRBSDV incidence (% plants infected) in NIP and OEP10‐12 plants. Error bars indicate ± standard deviation (SD).
Fig. S6 (A) Quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) results showing the expression levels of the Rice stripe virus (RSV) coat protein gene in RSV‐infected OEP10‐12 transgenic plants relative to the non‐transformed NIP controls at 30 days post‐inoculation (dpi). Results are shown for two different primer sets (CP1 and CP2). (B) RSV incidence (% plants infected) in NIP and OEP10‐12 plants. Error bars indicate ± standard deviation (SD).
Fig. S7 Scatterplot and KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis of differential gene expression in OEP10 transgenic plants. (A) Scatterplot analysis of differential gene expression in OEP10‐10 plants in contrast with the non‐transformed NIP controls. A red dot stands for one up‐regulated gene, a green dot for one down‐regulated gene and a blue dot for one non‐significantly changed gene. Genes were considered as being expressed and differentially regulated when they complied with the following criteria: false discovery rate (FDR) < 0.05 and the absolute value of log2(fold change) ratio > 1. Data were taken from three biological replicates. (B) TOP 10 pathway enrichment in OEP10‐10 plants in contrast with the controls.
Fig. S8 The effect of Rice stripe virus (RSV) on subsequent Rice black‐streaked dwarf virus (RBSDV) infection. (A) The relative expression levels of RBSDV genomic RNAs (S2, S5 and S10) in plants infected with RBSDV alone or jointly with RBSDV and RSV as assessed by quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) at 30 days post‐inoculation (dpi). (B) The relative expression levels of RSV coat protein (CP) gene in plants infected with RSV alone or jointly with RBSDV and RSV as assessed by RT‐qPCR at 30 dpi. Results are shown for two different primer sets (CP1 and CP2). Error bars indicate ± standard deviation (SD).
Table S1 The primers used in this study.
Table S2 Overview of small RNAs from mock‐ and Rice black‐streaked dwarf virus (RBSDV)‐infected plants.
Table S3 Transcriptome data obtained from OEP10-10 and control NIP plants.
Acknowledgements
The authors are indebted to Professor Yijun Zhou and Dr Ying Lan (Jiangsu Academy of Agricultural Sciences, China) for providing small brown planthoppers, to Professor Guohui Zhou (South China Agricultural University, China) for providing white‐backed planthoppers and to Professor Jianxiang Wu for providing RSV‐CP antibody. We thank Mike Adams for critical reading and improvement of the manuscript. This work was supported by grants from the National Key Research and Development Plan (2016YFD0200804), the State Basic Research Program of China (2014CB138403), the Major Project of New Varieties of Genetically Modified Organism of China (2014ZX0800104B), State Key Laboratory Breeding Base for Zhejiang Sustainable Pest and Disease Control (2010DS700124‐ZZ1801) and the National Natural Science Foundation of China (31601603). This work was sponsored by the K. C. Wong Magna Fund in Ningbo University. This work was also partially funded by the Scottish Government Rural and Environmental Science and Analytical Services Division (M.T. and S.M.).
References
- Abel, P. , Nelson, R. , De, B. , Hoffmann, N. , Rogers, S. , Fraley, R. and Beachy, R. (1986) Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science, 232, 738–743. [DOI] [PubMed] [Google Scholar]
- Akita, F. , Higashiura, A. , Shimizu, T. , Pu, Y. , Suzuki, M. , Uehara‐Ichiki, T. , Sasaya, T. , Kanamaru, S. , Arisaka, F. , Tsukihara, T. , Nakagawa, A. and Omura, T. (2012) Crystallographic analysis reveals octamerization of viroplasm matrix protein P9‐1 of Rice black streaked dwarf virus. J. Virol. 86, 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asurmendi, S. , Berg, R.H. , Smith, T.J. , Bendahmane, M. and Beachy, R.N. (2007) Aggregation of TMV CP plays a role in CP functions and in coat‐protein‐mediated resistance. Virology, 366, 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bance, V.B. (1991) Replication of potato virus X RNA is altered in coinfections with potato virus Y. Virology, 182, 486–494. [DOI] [PubMed] [Google Scholar]
- Beachy, R.N. , Loeschfries, S. and Tumer, N.E. (1990) Coat protein‐mediated resistance against virus infection. Annu. Rev. Phytopathol. 28, 451–472. [Google Scholar]
- Bendahmane, M. , Chen, I. , Asurmendi, S. , Bazzini, A.A. , Szecsi, J. and Beachy, R.N. (2007) Coat protein‐mediated resistance to TMV infection of Nicotiana tabacum involves multiple modes of interference by coat protein. Virology, 366, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capote, N. , Gorris, M.T. , Martínez, M.C. , Asensio, M. , Olmos, A. and Cambra, M. (2006) Interference between D and M types of Plum pox virus in Japanese plum assessed by specific monoclonal antibodies and quantitative real‐time reverse transcription‐polymerase chain reaction. Phytopathology, 96, 320–325. [DOI] [PubMed] [Google Scholar]
- Caracuel, Z. , Lozano‐Durán, R. , Huguet, S. , Arroyo‐Mateos, M. , Rodríguez‐Negrete, E.A. and Bejarano, E.R. (2012) C2 from Beet curly top virus promotes a cell environment suitable for efficient replication of geminiviruses, providing a novel mechanism of viral synergism. New Phytol. 194, 846–858. [DOI] [PubMed] [Google Scholar]
- Chávez‐Calvillo, G. , Contreras‐Paredes, C.A. , Mora‐Macias, J. , Noa‐Carrazana, J.C. , Serrano‐Rubio, A.A. , Dinkova, T.D. , Carrillo‐Tripp, M. and Silva‐Rosales, L. (2016) Antagonism or synergism between papaya ringspot virus and papaya mosaic virus in Carica papaya is determined by their order of infection. Virology, 489, 179–191. [DOI] [PubMed] [Google Scholar]
- Cheng, Z. , Li, S. , Gao, R. , Sun, F. , Liu, W. , Zhou, G. , Wu, J. , Zhou, X. and Zhou, Y . (2013) Distribution and genetic diversity of Southern rice black‐streaked dwarf virus in China. Virol J. 10, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S.‐Y. , Jeong, R.‐D. , Yoon, Y.‐N. , Lee, S.‐H. , Shin, D.B. , Kang, H.‐W. and Lee, B.C . (2013) One‐step multiplex reverse transcription‐polymerase chain reaction for the simultaneous detection of three rice viruses. J. Virol. Methods, 193, 674–678. [DOI] [PubMed] [Google Scholar]
- Fang, S. , Yu, J. , Feng, J. , Han, C. , Li, D. and Liu, Y. (2000) Identification of rice black‐streaked dwarf fijivirus in maize with rough dwarf disease in China. J. Agric. Biotechnol. 146, 167–170. [DOI] [PubMed] [Google Scholar]
- Folimonova, S.Y. (2013) Developing an understanding of cross‐protection by Citrus tristeza virus. Front. Microbiol. 4, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folimonova, S.Y. , Robertson, C.J. , Shilts, T. , Folimonov, A.S. , Hilf, M.E. , Garnsey, S.M. and Dawson, W.O . (2010) Infection with strains of Citrus tristeza virus does not exclude superinfection by other strains of the virus. J. Virol. 84, 1314–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa, N. , Itchoda, N. , Ishihara, T. , Goto, K. , Masuta, C. and Matsumura, T. (2010) HC‐Pro, a potyvirus RNA silencing suppressor, cancels cycling of Cucumber mosaic virus in Nicotiana benthamiana plants. Virus Genes, 40, 440–446. [DOI] [PubMed] [Google Scholar]
- García‐Cano, E. , Resende, R.O. , Fernández‐Muñoz, R. and Moriones, E. (2006) Synergistic interaction between Tomato chlorosis virus and Tomato spotted wilt virus results in breakdown of resistance in tomato. Phytopathology, 96, 1263–1269. [DOI] [PubMed] [Google Scholar]
- Germundsson, A. , Sandgren, M. , Barker, H. , Savenkov, E.I. and Valkonen, J.P.T. (2002) Initial infection of roots and leaves reveals different resistance phenotypes associated with coat protein gene‐mediated resistance to Potato mop‐top virus. J. Gen. Virol. 83, 1201–1209. [DOI] [PubMed] [Google Scholar]
- Gonsalves, D. (1998) Control of papaya ringspot virus in papaya: a case study. Annu. Rev. Phytopathol. 36, 415–437. [DOI] [PubMed] [Google Scholar]
- González‐Jara, P. , Tenllado, F. , Martínez‐García, B. , Atencio, F.A. , Barajas, D. , Vargas, M. , Diaz‐Ruiz, J. and Diaz‐Ruiz, J.R . (2004) Host‐dependent differences during synergistic infection by Potyviruses with potato virus X. Mol. Plant Pathol. 5, 29–35. [DOI] [PubMed] [Google Scholar]
- Haan, P.D. , Gielen, J.J.L. , Prins, M. , Wijkamp, I.G. , van Schepen, A. , Peters, D. , van Grinsven, M.Q.J.M. and Goldbach, R . (1992) Characterization of RNA‐mediated resistance to tomato spotted wilt virus in transgenic tobacco plants. Nat. Biotechnol. 10, 1133–1137. [DOI] [PubMed] [Google Scholar]
- He, Y. , Zhang, H. , Sun, Z. , Li, J. , Hong, G. , Zhu, Q. , Zhou, X. , MacFarlane, S. , Yan, F. and Chen, J. (2017) Jasmonic acid‐mediated defense suppresses brassinosteroid‐mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol. 214, 388–399. [DOI] [PubMed] [Google Scholar]
- Hisa, Y. , Suzuki, H. , Atsumi, G. , Choi, S.H. , Nakahara, K.S. and Uyeda, I. (2014) P3N‐PIPO of Clover yellow vein virus exacerbates symptoms in pea infected with White clover mosaic virus and is implicated in viral synergism. Virology, 449, 200–206. [DOI] [PubMed] [Google Scholar]
- Isogai, M. , Uyeda, I. and Lee, B.C. (1998) Detection and assignment of proteins encoded by rice black streaked dwarf fijivirus S7, S8, S9 and S10. J. Gen. Virol. 79, 1487–1494. [DOI] [PubMed] [Google Scholar]
- Kuribayashi, K. and Shinkai, A. (1952) On the new disease of rice, black‐streaked dwarf. Annu. Phytopathol. Soc. Jpn. 16, 41. [Google Scholar]
- Lan, Y. , Li, Y. , Zhiguo, E. , Sun, F. , Du, L. , Xu, Q. , Zhou, T. , Zhou, Y. and Fan, Y. (2017) Identification of virus‐derived siRNAs and their targets in RBSDV‐infected rice by deep sequencing. J. Basic Microb. 58, 227–237. [DOI] [PubMed] [Google Scholar]
- Latham, J.R. and Wilson, A.K. (2008) Transcomplementation and synergism in plants: implications for viral transgenes? Mol. Plant Pathol. 9, 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Andika, I.B. , Shen, J. , Lv, Y. , Ji, Y. , Sun, L. and Chen, J . (2013) Characterization of rice black‐streaked dwarf virus‐ and rice stripe virus‐derived siRNAs in singly and doubly infected insect vector Laodelphax striatellus . PLoS ONE, 8, e66007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Wang, H. and Zhou, G. (2014) Synergism between Southern rice black‐streaked dwarf virus and Rice ragged stunt virus enhances their insect vector acquisition. Phytopathology, 104, 794–799. [DOI] [PubMed] [Google Scholar]
- Li, S. , Wang, X. , Xu, J. , Ji, Y. and Zhou, Y. (2015) A simplified method for simultaneous detection of Rice stripe virus and Rice black‐streaked dwarf virus in insect vector. J. Virol. Methods, 211, 32–35. [DOI] [PubMed] [Google Scholar]
- Lindbo, J.A. and Falk, B.W. (2017) The impact of "coat protein‐mediated virus resistance" in applied plant pathology and basic research. Phytopathology, 107, 624–634. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Wei, C. , Zhong, Y. and Li, Y. (2007a) Rice black‐streaked dwarf virus minor core protein P8 is a nuclear dimeric protein and represses transcription in tobacco protoplasts. Febs Lett. 581, 2534–2540. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Wei, C. , Zhong, Y. and Li, Y. (2007b) Rice black‐streaked dwarf virus outer capsid protein P10 has self‐interactions and forms oligomeric complexes in solution. Virus Res. 127, 34–42. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Pu, L. , Xie, G. , Ji, C. , Ling, B. , Zhang, M. , Xu, D. and Zhou, G . (2012) Transmission characteristics of Southern rice black‐streaked dwarf virus by rice planthoppers. Crop Prot. 41, 71–76. [Google Scholar]
- Ratcliff, F.G. , MacFarlane, S.A. and Baulcombe, D.C. (1999) Gene silencing without DNA: RNA‐mediated cross‐protection between viruses. Plant Cell, 11, 1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata, E. and Kitagawa, Y. (1977) Rice black‐streaked dwarf virus: its properties, morphology and intracellular localization. Virology, 77, 826–842. [DOI] [PubMed] [Google Scholar]
- Sun, L. , Xie, L. , Andika, I.B. , Tan, Z. and Chen, J. (2013) Non‐structural protein P6 encoded by rice black‐streaked dwarf virus is recruited to viral inclusion bodies by binding to the viroplasm matrix protein P9–1. J. Gen. Virol. 94, 1908–1916. [DOI] [PubMed] [Google Scholar]
- Sun, Z. , He, Y. , Li, J. , Wang, X. and Chen, J. (2015) Genome‐wide characterization of rice black streaked dwarf virus‐responsive microRNAs in rice leaves and roots by small RNA and degradome sequencing. Plant Cell Physiol. 56, 688–699. [DOI] [PubMed] [Google Scholar]
- Sun, Z. , Yang, D. , Xie, L. , Sun, L. , Zhang, S. , Zhu, Q. , Li, J. , Wang, X. and Chen, J. (2013a) Rice black‐streaked dwarf virus P10 induces membranous structures at the ER and elicits the unfolded protein response in Nicotiana benthamiana . Virology, 447, 131–139. [DOI] [PubMed] [Google Scholar]
- Sun, Z. , Zhang, S. , Xie, L. , Zhu, Q. , Tan, Z. , Bian, J. , Sun, L. and Chen, J. (2013b) The secretory pathway and the actomyosin motility system are required for plasmodesmatal localization of the P7–1 of rice black‐streaked dwarf virus. Arch. Virol. 158, 1055–1064. [DOI] [PubMed] [Google Scholar]
- Syller, J. (2012) Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 13, 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syller, J. and Grupa, A. (2016) Antagonistic within‐host interactions between plant viruses: molecular basis and impact on viral and host fitness. Mol. Plant Pathol. 17, 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, T. , Zhou, C.J. , Wang, Q. , Chen, X.R. , Sun, Q. , Zhao, T.Y. , Ye, J.‐C. , Wang, Y. , Zhang, Z.‐Y. , Zhang, Y.‐L. , Guo, Z.‐J. , Wang, X.‐B. , Li, D.‐W. , Yu, J.‐L. and Han, C.‐G . (2017) Rice black streaked dwarf virus P7–2 forms a SCF complex through binding to Oryza sativa SKP1‐like proteins, and interacts with GID2 involved in the gibberellin pathway. PLoS ONE, 12, e0177518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatineni, S. and French, R. (2016) The coat protein and NIa protease of two Potyviridae family members independently confer superinfection exclusion. J. Virol. 90, 10 886–10 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untiveros, M. , Fuentes, S. and Salazar, L.F. (2007) Synergistic interaction of Sweet potato chlorotic stunt virus (Crinivirus) with carla‐, cucumo‐, ipomo‐, and potyviruses infecting sweet potato. Plant Dis. 91, 669–676. [DOI] [PubMed] [Google Scholar]
- Valkonen, J.P. , Rajamäki, M.L. and Kekarainen, T. (2002) Mapping of viral genomic regions important in cross‐protection between strains of a potyvirus. Mol. Plant–Microbe Interact. 15, 683–692. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Liu, Y. , He, J. , Zheng, X. , Hu, J. , Liu, Y. , Dai, H. , Zhang, Y. , Wang, B. , Wu, W. , Gao, H. , Zhang, Y. , Tao, X. , Deng, H. , Yuan, D. , Jiang, L. , Zhang, X. , Guo, X. , Cheng, X. , Wu, C. , Wang, H. , Yuan, L. and Wan, J . (2014) STV11 encodes a sulphotransferase and confers durable resistance to rice stripe virus. Nat. Commun. 5, 4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Tao, T. , Zhang, Y. , Wu, W. , Li, D. , Yu, J. and Han, C . (2011) Rice black‐streaked dwarf virus P6 self‐interacts to form punctate, viroplasm‐like structures in the cytoplasm and recruits viroplasm‐associated protein P9–1. Virol. J. 8, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.H. , Fang, S.G. , Xu, J.L. , Sun, L.Y. , Li, D.W. and Yu, J.L. (2003) Sequence analysis of the complete genome of rice black‐streaked dwarf virus isolated from maize with rough dwarf disease. Virus Genes, 27, 163–168. [DOI] [PubMed] [Google Scholar]
- Wei, T. and Li, Y. (2016) Rice reoviruses in insect vectors. Annu. Rev. Phytopathol. 54, 99–120. [DOI] [PubMed] [Google Scholar]
- Wei, T. , Yang, J. , Liao, F. , Gao, F. , Lu, L. , Zhang, X. , Li, F. , Wu, Z‐J , Lin, Q‐Y , Xie, L‐H and Lin, H‐X . (2009) Genetic diversity and population structure of rice stripe virus in China. J. Gen. Virol. 90, 1025–1034. [DOI] [PubMed] [Google Scholar]
- Wu, J. , Ni, Y. , Liu, H. , Rao, L. , Zhou, Y. and Zhou, X. (2013) Development and use of three monoclonal antibodies for the detection of rice black‐streaked dwarf virus in field plants and planthopper vectors. Virol. J. 10, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, L. , Lv, M.F. , Song, X.J. , Yang, J. , Li, J. , Sun, Z.T. , Zhang, Y.‐Y. , Chen, J.‐P. and Zhang, H.‐M. . (2017) Structure and components of the globular and filamentous viroplasms induced by Rice black‐streaked dwarf virus. Micron, 98, 12–23. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Chen, J. , Lei, J. and Adams, M.J. (2001) Sequence analysis shows that a dwarfing disease on rice, wheat and maize in China is caused by rice black‐streaked dwarf virus. Eur. J. Plant Pathol. 107, 563–567. [Google Scholar]
- Zhang, X.‐F. , Sun, R. , Guo, Q. , Zhang, S. , Meulia, T. , Halfmann, R. , Li, D. and Qu, F . (2017) A self‐perpetuating repressive state of a viral replication protein blocks superinfection by the same virus. Plos Pathog. 13, e1006253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, G. , Xu, D. and Zhang, M. (2013) Southern rice black‐streaked dwarf virus: a white‐backed planthopper‐transmitted fijivirus threatening rice production in Asia. Front. Microbiol. 4, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Yang, Y. , Wang, X. , Yu, F. , Yu, C. , Chen, J. , Cheng, Y. , Yan, C. and Chen, J. (2013) Enhanced transgene expression in rice following selection controlled by weak promoters. Bmc Biotechnol. 13, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, T. , Wu, L.J. , Wang, Y. , Cheng, Z.B. , Ji, Y.H. , Fan, Y.J. and Zhou, Y. (2010) Preliminary report on the transmission of rice black‐streaked dwarf virus from frozen infected leaves to rice by insect vector small brown planthopper (Laodelphax striatellus). Chin. J. Rice Sci. 24, 425–428. [Google Scholar]
- Ziebell, H. and Carr, J.P. (2009) Effects of dicer‐like endoribonucleases 2 and 4 on infection of Arabidopsis thaliana by cucumber mosaic virus and a mutant virus lacking the 2b counter‐defence protein gene. J. Gen. Virol. 90, 2288–2292. [DOI] [PubMed] [Google Scholar]
- Ziebell, H. and Carr, J.P. (2010) Cross‐protection: a century of mystery. Adv. Virus Res. 76, 211–264. [DOI] [PubMed] [Google Scholar]
- Ziebell, H. , Payne, T. , Berry, J.O. , Walsh, J.A. and Carr, J.P. (2007) A cucumber mosaic virus mutant lacking the 2b counter‐defence protein gene provides protection against wild‐type strains. J. Gen. Virol. 88, 2862–2871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Western blotting to detect P10 protein expression in OEP10 and P10RNA transgenic plants.
Fig. S2 The height of mock‐ and Rice black‐streaked dwarf virus (RBSDV)‐infected (+RB) NIP, OEP10‐10 and OEP10‐12 plants. Error bars indicate ± standard deviation (SD).
Fig. S3 The relative expression levels of some Rice black‐streaked dwarf virus (RBSDV) RNAs in RBSDV‐infected NIP, OEP10‐10 and OEP10‐12 plants. RNA levels of S4, S5, S7 and S10 were tested by quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) at different times in RBSDV‐infected plants. Error bars indicate ± standard deviation (SD). An asterisk at the top of a column indicates a significant difference at P < 0.05.
Fig. S4 (A) Quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) results showing the expression levels of Rice black‐streaked dwarf virus (RBSDV) RNA segments S5, S6 and S8 in RBSDV‐infected P10 RNA transgenic plants relative to the non‐transformed NIP controls at 30 days post‐inoculation (dpi). (B) RBSDV incidence (% plants infected) in NIPand P10 RNA plants. Error bars indicate ± standard deviation (SD).
Fig. S5 (A) Quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) results showing the expression levels of Southern rice black‐streaked dwarf virus (SRBSDV) segments S1–S10 in SRBSDV‐infected OEP10‐12 transgenic plants relative to the non‐transformed NIP controls at 30 days post‐inoculation (dpi). (B) SRBSDV incidence (% plants infected) in NIP and OEP10‐12 plants. Error bars indicate ± standard deviation (SD).
Fig. S6 (A) Quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) results showing the expression levels of the Rice stripe virus (RSV) coat protein gene in RSV‐infected OEP10‐12 transgenic plants relative to the non‐transformed NIP controls at 30 days post‐inoculation (dpi). Results are shown for two different primer sets (CP1 and CP2). (B) RSV incidence (% plants infected) in NIP and OEP10‐12 plants. Error bars indicate ± standard deviation (SD).
Fig. S7 Scatterplot and KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis of differential gene expression in OEP10 transgenic plants. (A) Scatterplot analysis of differential gene expression in OEP10‐10 plants in contrast with the non‐transformed NIP controls. A red dot stands for one up‐regulated gene, a green dot for one down‐regulated gene and a blue dot for one non‐significantly changed gene. Genes were considered as being expressed and differentially regulated when they complied with the following criteria: false discovery rate (FDR) < 0.05 and the absolute value of log2(fold change) ratio > 1. Data were taken from three biological replicates. (B) TOP 10 pathway enrichment in OEP10‐10 plants in contrast with the controls.
Fig. S8 The effect of Rice stripe virus (RSV) on subsequent Rice black‐streaked dwarf virus (RBSDV) infection. (A) The relative expression levels of RBSDV genomic RNAs (S2, S5 and S10) in plants infected with RBSDV alone or jointly with RBSDV and RSV as assessed by quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) at 30 days post‐inoculation (dpi). (B) The relative expression levels of RSV coat protein (CP) gene in plants infected with RSV alone or jointly with RBSDV and RSV as assessed by RT‐qPCR at 30 dpi. Results are shown for two different primer sets (CP1 and CP2). Error bars indicate ± standard deviation (SD).
Table S1 The primers used in this study.
Table S2 Overview of small RNAs from mock‐ and Rice black‐streaked dwarf virus (RBSDV)‐infected plants.
Table S3 Transcriptome data obtained from OEP10-10 and control NIP plants.


