FIGURE 3.
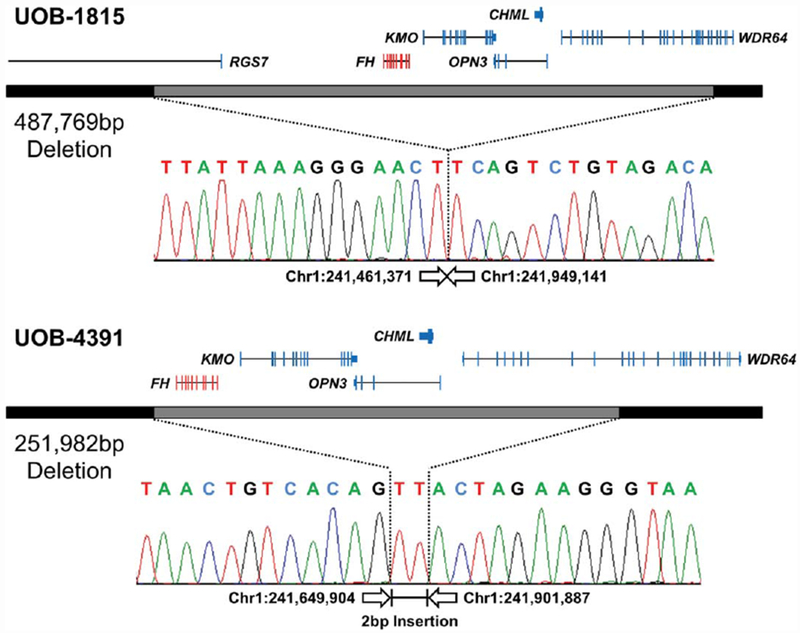
Breakpoint mapping of two CGH array-based FH complete deletions in HLRCC patients. The Agilent custom high-definition CGH array refined the breakpoints down to the region between the first and last probes that demonstrated no loss or 50% loss. A combination of primers was designed to sit within the potentially retained regions near each end of the deletion and used to PCR amplify across the deleted region. Sequencing of these PCR products demonstrated the breakpoint at the nucleotide level for two deletions with one containing a 2 bp insertion. The completely or partially lost genes are indicated above the deletion with the target FH gene in red and the additionally deleted genes in blue. All positions and size calculations are based on the GRCh37/hg19 build. [Color figure can be viewed at wileyonlinelibrary.com]
