Figure 4.
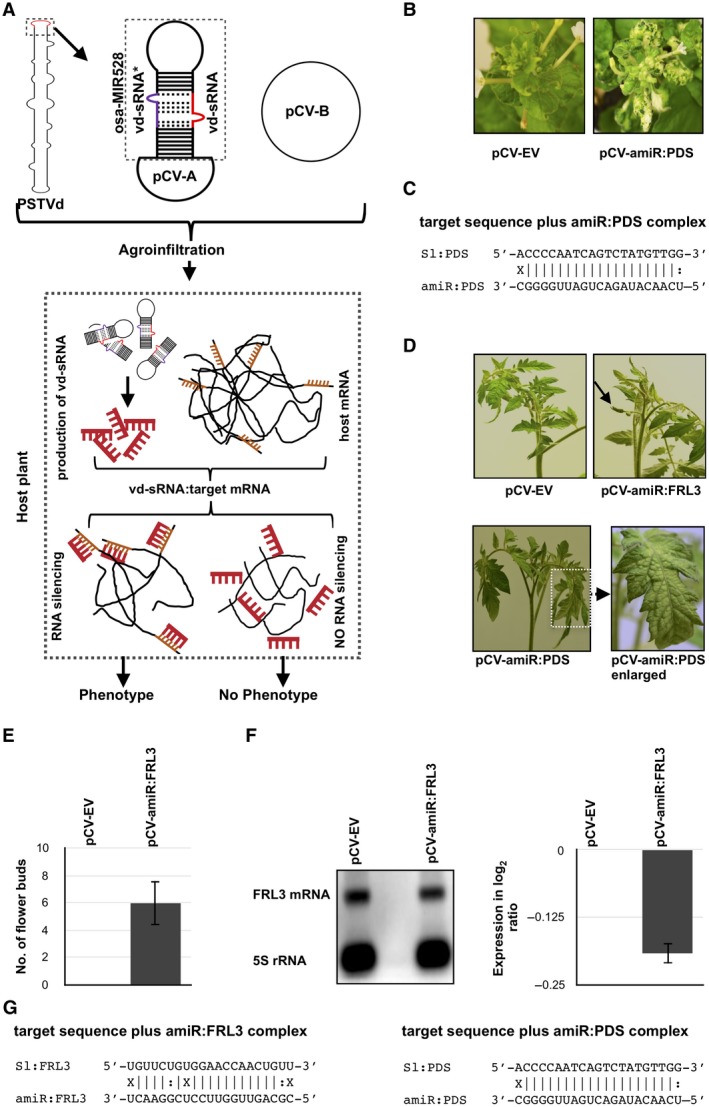
Transient expression of viroid‐derived small RNA (vd‐sRNA) induces phenotypic changes in tomato cv. Rutgers. (A) Flow chart illustrating the details of the vd‐sRNA:virus‐induced gene silencing (VIGS) experiment. The vd‐sRNA expressed as an artificial microRNA (amiRNA) in the pCV‐A vector was agroinfiltrated, together with pCV‐B, into plants at the cotyledon stage. The specific interaction of vd‐sRNA with the target sequence leads to either RNA‐induced silencing complex (RISC)‐mediated cleavage or translational repression, which, in turn, results in the phenotype of the plant. (B) The vd‐sRNA:VIGS assay was verified by agroinfiltration of pCV‐amiR:PDS into Nicotiana benthamiana plants. The resulting plants showed bleaching at 35 days post‐infection (dpi). (C) Duplexes predicted to be formed by the complexes of vd‐sRNAs and their target sequences. (D) The tomato plants were subjected to a vd‐sRNA:VIGS assay in order to evaluate the ability of the vd‐sRNA to induce early flowering. At 35 dpi, plants agroinfiltrated with pCV‐amiR:FRL3 exhibited early flower buds. pCV‐EV, plants agroinfiltrated with pCV emply vector; pCV‐amiR:PDS, plants agroinfiltrated with pCV‐amiR:PDS, which is capable of inducing the RNA silencing of the phytoene desaturase (PDS) mRNA; pCV‐amiR:FRL3, plants agroinfiltrated with pCV‐amiR:FRL3, which is capable of producing sRNA similar to the vd‐sRNA that was predicted to bind to FRIGIDA‐like protein 3 (FRL3) mRNA. (E) The total number of flowers observed in three separate experiments were recorded. The error bars indicate the standard deviation (SD). (F) Total RNA extracted from systemic leaf samples from the agroinfiltrated plants was subjected to an RNA gel blot assay in order to evaluate the effect of vd‐sRNA on the FRL3 mRNA using FRL3‐specific radiolabelled probes. The resulting gel blot signals (from the right panel) were quantified and expressed as a log2 ratio of FRL3 mRNA to the 5S rRNA signal. (G) Duplexes predicted to be formed by complexes of vd‐sRNA and their target sequences. [Colour figure can be viewed at wileyonlinelibrary.com]
