Figure 4.
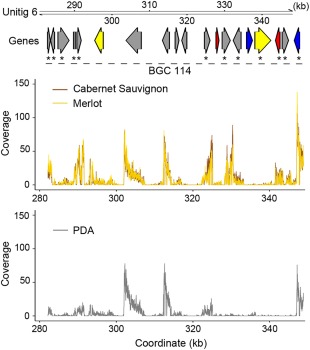
Example of Neofusicoccum parvum gene clusters modulated as a function of in vitro substrate. Arrows represent genes coding for carbohydrate‐active enzymes (CAZymes) (red), P450s (blue) and transporters (yellow). Grey arrows correspond to genes predicted to be part of a secondary metabolism cluster, but with other annotations. Asterisks highlight genes with significant up‐regulation in the presence of wood in vitro. Line plots show the RNA‐sequencing mapping coverage in samples grown on minimal medium amended with finely ground grape wood of either ‘Cabernet Sauvignon’ or ‘Merlot’ or on potato dextrose agar (PDA) without wood.
