Figure 1.
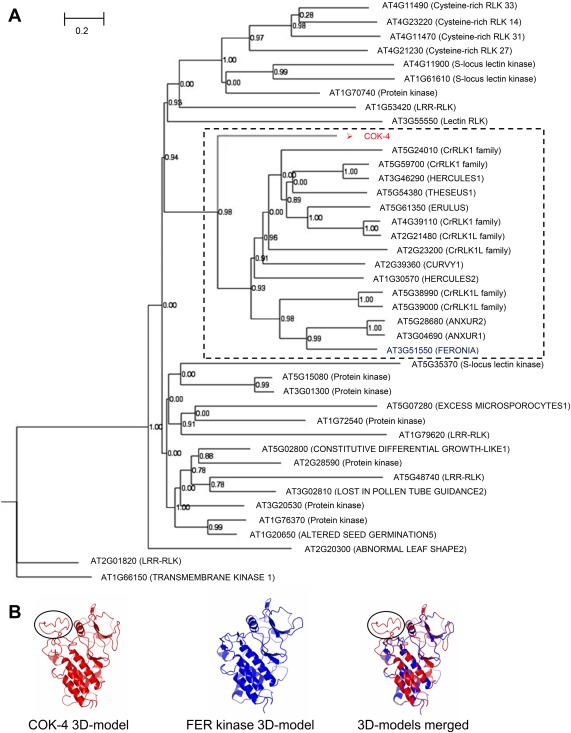
COK‐4 is highly similar to the Arabidopsis FERONIA (FER) kinase domain. (A) Phylogenetic tree showing the clustering of the predicted COK‐4 protein with 15 of the 17 Catharanthus roseus RLK1‐like (CrRLK1L) family members of Arabidopsis (broken square). The phylogenetic tree was obtained using the top 40 different protein kinases of Arabidopsis with significant alignment (threshold E‐value ≤ 3 × 10−33) with the predicted COK‐4 protein from the common bean line SEL 1308 [National Center for Biotechnology Information (NCBI) accession number AAF98554; Melotto and Kelly, 2001], using the maximum parsimony method from MAFFT software (Katoh and Standley, 2013). The nodes were confirmed by 1000 bootstraps. (B) Three‐dimensional (3D) protein structure of FER and COK‐4 kinase domains showing that both kinases have similar structures. The protein models were obtained with SWISS‐MODEL (https://swissmodel.expasy.org) and the alignment of the models was made using CCP4G v.2.10.6 (McNicholas et al., 2011). The black circles indicate the COK‐4 protein region that does not fit within the FER kinase domain structure. This region contains the transmembrane region of COK‐4 predicted by Melotto and Kelly (2001). LRR, leucine‐rich repeat; RLK, receptor‐like kinase.
