Figure 1.
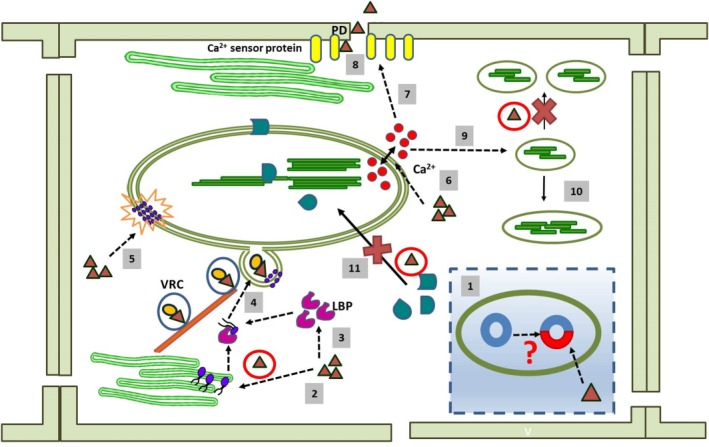
Conceptual depiction of different aspects of chloroplast–virus interaction. For simplified representation, the same symbols have been used for different viruses (filled red triangles). During the course of evolution, viruses have probably replaced some of the genes in the chloroplast with their own genetic material; this might be reflected by the plastid affinity of viruses (1). Viruses alter the lipid biosynthesis (lipid molecules are indicated by blue circles) (2) and lipid trafficking pathway by the overexpression of lipid‐binding proteins (LBPs) (3). These alterations aid in membrane invagination, vesicle formation (4) and rearrangement of the membrane lipid bilayer (5). Viral infection affects Ca2+ (Ca2+ ions indicated by red circles) signalling mediated by the chloroplast (6). This might affect the biochemical properties of synaptotagmin (SYTA)‐like Ca2+ sensor proteins (7) which, in turn, help the viral component to move through the plasmodesmata (PD) for cell‐to‐cell movement (8). A change in the Ca2+ level also hampers normal chloroplast division (9), and large, abnormally shaped chloroplasts are formed (10). Viral proteins sequester various chloroplast‐localized proteins (chloroplast‐localized proteins are indicated by green geometric shapes) in the cytosol (11); this again damages the organelle structurally and functionally. VRC, virus replication complex.
