Figure 2.
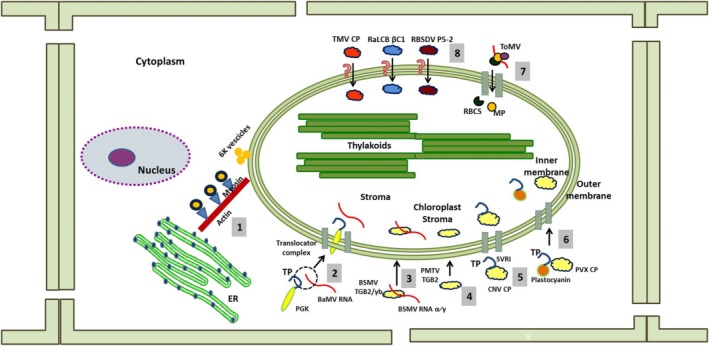
Different strategies adopted by different viruses to send their nucleic acid and/or protein products into the chloroplast. Vesicles induced by the 6K protein of Turnip mosaic virus (TuMV) follow the endoplasmic reticulum (ER)–Golgi vesicular transport pathway to reach the chloroplast using the actomyosin motility system (1). The 3′ untranslated region (UTR) of Bamboo mosaic virus (BaMV) RNA binds with the transit peptide of p51, a chloroplast phosphoglycerate kinase (chl‐PGK), to pass through the membranes of the chloroplast (2). TGB2, the movement protein of Barley stripe mosaic virus (BSMV) and Potato mop‐top virus (PMTV), shows different properties when localized inside the chloroplast in terms of being associated with or without the respective nucleic acids (3, 4). The arm domain of Cucumber necrosis virus (CNV) coat protein (CP) has an embedded sequence identical to the transit peptide (TP) of chloroplast proteins, which helps the viral protein to cross the translocation machinery (5). The Potato virus X (PVX) CP interacts with the TP of plastocyanine to reach the organelle (6). The movement protein (MP) of Tomato mosaic virus (ToMV) interacts with the small subunit of RuBisCO and localizes to the chloroplast (7). The mechanisms used by a few proteins, such as Tobacco mosaic virus (TMV) CP, Radish leaf curl betasatellite (RaLCB) βC1 and Rice black‐streaked dwarf virus (RBSDV) P5‐2, to pass through the translocation machinery are yet not known (8). RBCS, small subunit of RuBisCO; SVRI, R‐arm region plus the first 4 aa [SVRI] of the shell [S] domain of CNV coat protein.
