Summary
Fusarium graminearum is a causal agent of wheat scab and a producer of the trichothecene mycotoxin deoxynivalenol (DON). The expression of trichothecene biosynthesis (TRI) genes and DON production are mainly regulated by the cyclic adenosine monophosphate‐protein kinase A (cAMP‐PKA) pathway and two pathway‐specific transcription factors (TRI6 and TRI10). Interestingly, deletion mutants of TRI6 show reduced expression of several components of cAMP signalling, including the FgCAP1 adenylate‐binding protein gene that has not been functionally characterized in F. graminearum. In this study, we show that FgCap1 interacts with Fac1 adenylate cyclase and that deletion of FgCAP1 reduces the intracellular cAMP level and PKA activity. The Fgcap1 deletion mutant is defective in vegetative growth, conidiogenesis and plant infection. It also shows significantly reduced DON production and TRI gene expression, which can be suppressed by exogenous cAMP, indicating a PKA‐dependent regulation of DON biosynthesis by FgCap1. The wild‐type, but not tri6 mutant, shows increased levels of intracellular cAMP and FgCAP1 expression under DON‐producing conditions. Furthermore, the promoter of FgCAP1 contains one putative Tri6‐binding site that is important for its function during DON biosynthesis, but is dispensable for hyphal growth, conidiogenesis and pathogenesis. In addition, FgCap1 shows an actin‐like localization to the cortical patches at the apical region of hyphal tips. Phosphorylation of FgCap1 at S353 was identified by phosphoproteomics analysis. The S353A mutation in FgCAP1 has no effect on its functions during vegetative growth, conidiation and DON production. However, expression of the FgCAP1 S353A allele fails to complement the defects of the Fgcap1 mutant in plant infection, indicating the importance of the phosphorylation of FgCap1 at S353 during pathogenesis. Taken together, our results suggest that FgCAP1 is involved in the regulation of DON production via cAMP signalling and subjected to a feedback regulation by TRI6, but the phosphorylation of FgCap1 at S353 is probably unrelated to the cAMP‐PKA pathway because the S353A mutation only affects plant infection.
Keywords: cAMP signalling, F‐actin, feedback regulation, mycotoxin, pathogenic growth, phosphorylation
Introduction
Fusarium head blight (FHB), caused by Fusarium graminearum, is one of the most destructive diseases of wheat and barley in many wheat‐growing regions (Goswami and Kistler, 2004). In addition to causing severe yield losses, F. graminearum produces the trichothecene mycotoxin deoxynivalenol (DON) in infested grains. As a potent inhibitor of protein synthesis in eukaryotic organisms, DON is harmful to human and animal health (Audenaert et al., 2014; Van de Walle et al., 2010). DON is also an important virulence factor during plant infection. Although it is not required for F. graminearum to establish initial infection, DON plays a critical role in the spread from infected kernels to neighbouring spikelets (Bai et al., 2002). Deletion mutants of the TRI5 gene, encoding the trichodiene synthase that catalyses the initial step of DON biosynthesis, can cause diseases on inoculated kernels, but fail to spread infection to other spikelets on the same wheat heads. The expression of TRI5 is induced in initial infection structures, further indicating the importance of DON in plant infection (Boenisch and Schafer, 2011).
In F. graminearum, most of the genes involved in trichothecene biosynthesis and transport (TRI genes), including TRI4, TRI5, TRI6, TRI10 and TRI12, are located in the main TRI gene cluster (Brown et al., 2004). TRI4, like TRI1, encodes a cytochrome P‐450 oxygenase that localizes to toxiosomes, distinct vesicles specific for DON biosynthesis (Menke et al., 2013). The Tri12 major facilitator superfamily (MFS) transporter interacts with toxiosomes for the efflux of trichothecene mycotoxins (Menke et al., 2012). The two transcriptional factor genes in the cluster, TRI6 and TRI10, are important for the regulation of TRI gene expression (Seong et al., 2009). tri6 and tri10 mutants are defective in DON production and in the expression of TRI genes under DON‐producing conditions (Seong et al., 2009). Although the Tri10‐binding site has not been identified, the putative Tri6‐binding sites identified in three different studies are different, probably as a result of the different approaches used (Hohn et al., 1999; Nasmith et al., 2011; Seong et al., 2009; Sieber et al., 2014). Two studies were based on bioinformatics analyses with microarray data (Seong et al., 2009; Sieber et al., 2014), but the other used chromatin immunoprecipitation sequencing (ChIP‐seq) and gel‐mobility shift assays (Nasmith et al., 2011). It is also possible that Tri6 and Tri10 can form homo‐ and hetero‐dimers that differ in the recognition of DNA sequences.
Various environmental factors or physiological conditions are known to affect DON biosynthesis in the wheat scab fungus, possibly by the activation of Tri6 and/or Tri10 to regulate TRI gene expression (Hou et al., 2015; Jiang et al., 2015; Merhej et al., 2011). As in many other mycotoxin‐producing fungi (Roze et al., 2004; Schmidt‐Heydt et al., 2015), the cyclic adenosine monophosphate‐protein kinase A (cAMP‐PKA) signal transduction pathway plays a regulatory role in DON biosynthesis in F. graminearum (Bormann et al., 2014; Guo et al., 2016; Hu et al., 2014; Jiang et al., 2016b). Exogenous cAMP treatment stimulates TRI gene expression and cellular differentiation associated with DON production (Jiang et al., 2016b). Functional characterizations of the two genes encoding the catalytic subunits of PKA have shown that CPK1 plays a major role in the regulation of DON production, although CPK2 also contributes (Hu et al., 2014). The cpk1 cpk2 double mutant, but not the cpk1 or cpk2 mutant, is completely blocked in DON biosynthesis. Although the Pde1 and Pde2 cAMP phosphodiesterases negatively regulate DON production in F. graminearum (Jiang et al., 2016b), the FAC1 adenylate cyclase responsible for the synthesis of intracellular cAMP is essential for TRI gene expression and trichothecene biosynthesis (Bormann et al., 2014; Hu et al., 2014). In addition, a gain‐of‐function mutation in the adenylyl cyclase gene results in the overproduction of DON under repressive conditions in F. graminearum (Blum et al., 2016).
In the budding yeast Saccharomyces cerevisiae, the SRV2 adenylate cyclase‐associated protein (CAP) gene has been identified as a suppressor of a constitutively active RAS2 G19V mutation (Fedor‐Chaiken et al., 1990). SRV2 has been shown to be required for RAS‐mediated activation of adenylate cyclase and null mutations in SRV2 are lethal in yeast (Fedor‐Chaiken et al., 1990; Yu et al., 1999). The N‐terminal region of Srv2 has an adenylyl cyclase‐binding domain (ACB) that is responsible for its functional relationship with RAS (Shi et al., 1997). Srv2 also has a C‐terminal actin‐binding domain (ABD) that is important for its functions in actin turnover and polarized growth (Bertling et al., 2004; Quintero‐Monzon et al., 2009; Zhou et al., 2012). These two domains are well conserved in SRV2 and its orthologues from other fungi. Studies in a few fungal pathogens have shown that CAP proteins are generally important for the activation of adenylate cyclase and cAMP signalling (Bahn and Sundstrom, 2001; Fedor‐Chaiken et al., 1990; Zhou et al., 2012). ACA1, the SRV2 orthologue in the human pathogen Cryptococcus neoformans, is important for glucose‐induced cAMP accumulation, mating and capsule production (Bahn et al., 2004). In another human pathogen, Candida albicans, CAP1‐mediated cAMP signalling is required for the transition of yeast cells to hyphal growth (Bahn and Sundstrom, 2001). In the plant‐pathogenic fungi Ustilago maydis and Magnaporthe oryzae, CAP proteins are important for infection‐related morphogenesis and pathogenesis (Takach and Gold, 2010; Zhou et al., 2012). As in S. cerevisiae, deletion of the CAP1 gene suppresses the effects of a dominant RAS2 mutation in M. oryzae (Zhou et al., 2012).
In F. graminearum, the Tri6 transcription factor has been shown to play a critical role in the regulation of DON biosynthesis by the cAMP‐PKA pathway (Hu et al., 2014; Jiang et al., 2016b). Interestingly, microarray analysis with the tri6 mutant (Seong et al., 2009) has shown that deletion of TRI6 reduces the expression of several key components of the PKA pathway under DON‐inducing conditions, including FAC1, CPK1, CPK2 and FGSG_01923 (an orthologue of yeast SRV2 and M. oryzae CAP1), indicating a possible feedback regulation of cAMP signalling by Tri6 during DON production. Unlike other genes related to cAMP signalling, FGSG_01923 (named FgCAP1 in this study) has not been functionally characterized in F. graminearum. In this study, we show that FgCAP1 is important for vegetative growth, conidiogenesis, plant infection and DON production. As an Fac1‐interacting protein, FgCap1 is required for the maintenance of a normal intracellular cAMP level to properly regulate PKA activity. However, phosphorylation of FgCap1 at S353 is important for plant infection, but dispensable for DON biosynthesis. Overall, our results indicate that FgCAP1 is involved in the regulation of DON production via cAMP signalling and is subjected to a feedback regulation by TRI6, but phosphorylation‐dependent functions of FgCap1 during plant infection may not be related to cAMP signalling.
Results
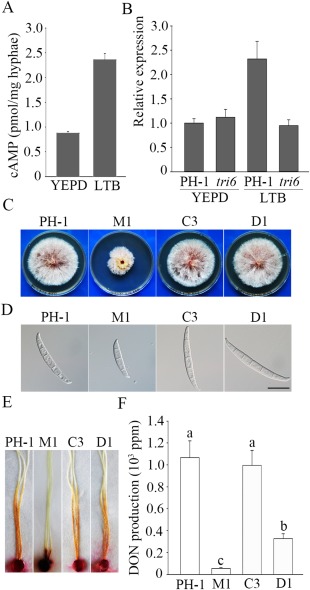
FgCAP1 is important for vegetative growth and conidium morphology
FGSG_01923 (named FgCAP1 in this study) is the distinct orthologue of S. cerevisiae SRV2 and M. oryzae CAP1. It has the typical structural components of CAP proteins, including an ACB and an ABD. To determine its function in DON production and cAMP signalling, we generated the Fgcap1 mutant M1 in the wild‐type strain PH‐1 with the split‐marker approach (Catlett et al., 2003). In comparison with PH‐1, the Fgcap1 mutant showed a reduced growth rate (Fig. 1A; Table 1). Mutant colonies tended to have irregular edges and produced more compact aerial hyphae than the wild‐type. When cultured at an elevated temperature (30 ºC), the wild‐type formed smaller colonies in comparison with cultures incubated at 25 ºC. For the Fgcap1 mutant, the growth rate was similar between cultures grown at 30 or 25 ºC (Fig. 1A), indicating that the Fgcap1 mutant shows increased tolerance to high temperature.
Figure 1.
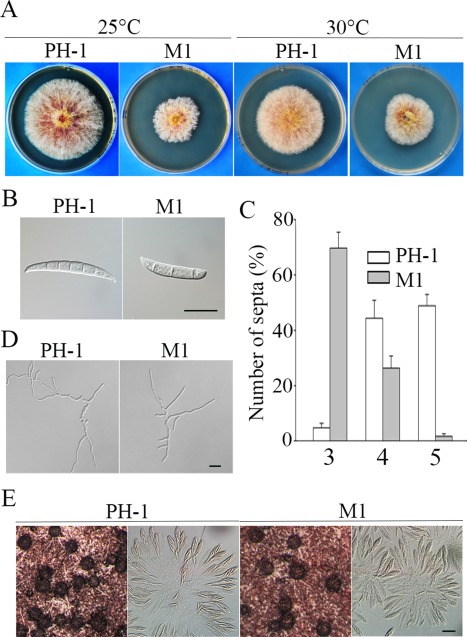
Phenotypes of the Fgcap1 mutant in vegetative growth, conidiogenesis, germination and sexual reproduction. (A) Three‐day‐old potato dextrose agar (PDA) cultures of the wild‐type (PH‐1) and Fgcap1 mutant (M1) strains. (B) Conidium morphology of PH‐1 and M1. Bar, 20 μm. (C) Average number of septa in conidia of PH‐1 and mutant M1 harvested from 5‐day‐old carboxymethyl cellulose (CMC) cultures. (D) Germlings of PH‐1 and M1 after incubation in yeast extract peptone dextrose (YEPD) medium for 10 h. Bar, 20 μm. (E) Perithecia and asci of PH‐1 and M1 formed on selfing cultures 2 weeks after induction. Bar, 20 μm.
Table 1.
Defects of Fgcap1 mutant in growth, conidiation, deoxynivalenol (DON) production and virulence.
| Strain | Growth rate (mm/day)* | Conidiation (105 conidia/mL) † | DON production (ppm) ‡ | Disease index § | |
|---|---|---|---|---|---|
| Rice grains | Wheat kernels | ||||
| PH‐1 | 11.8 ± 0.3A | 10.5 ± 0.3A | 922.3 ± 55.0A | 506.2 ± 42.2A | 13.2 ± 2.6A |
| M1 | 6.1 ± 0.1B | 10.6 ± 0.3A | 75.9 ± 16.4B | 22.9 ± 18.1B | 0.7 ± 0.5B |
| C3 | 12.0 ± 0.1A | 10.3 ± 0.6A | 874.6 ± 107.1A | 525.0 ± 26.4A | 12.9 ± 1.8A |
Means and standard deviations were calculated using the results from three replicates for growth rate and conidiation assays and from five replicates for DON and infection assays. Data were analysed with Duncan's pairwise comparison. Different letters mark statistically significant differences (P = 0.05).
*Average daily extension in colony radius on potato dextrose agar (PDA) plates.
†Conidiation in 5‐day‐old carboxymethyl cellulose (CMC) cultures.
‡DON production assayed with rice grain cultures and diseased wheat kernels from symptomatic spikelets.
§Disease index was estimated as the number of diseased spikelets on each inoculated wheat head at 14 days post‐inoculation (dpi).
In 5‐day‐old CMC cultures, conidiation appeared to be normal (Table 1), but conidia produced by the Fgcap1 mutant were shorter and had fewer septa than those of PH‐1 (Fig. 1B). Approximately 69.6% of the Fgcap1 conidia had only three septa, whereas only 4.8% of the wild‐type conidia had fewer than four septa (Fig. 1C). Nevertheless, Fgcap1 conidia with fewer septa showed normal germination, although germ tube growth was significantly reduced (Fig. 1D). On selfing plates, the Fgcap1 mutant produced abundant perithecia with normal morphology and size. No obvious defects in perithecium development and ascospore development were observed (Fig. 1E). These results indicate that FgCAP1 is dispensable for sexual reproduction, although it is important for hyphal growth and conidiogenesis.
Fgcap1 mutant is defective in plant infection
In infection assays with wheat heads, unlike the wild‐type that spread from the inoculation site to other spikelets, the Fgcap1 mutant only caused FHB symptoms on the inoculated spikelets (Fig. 2A). On average, the disease index of the Fgcap1 mutant was 0.7 (Table 1). The average disease index was 13.2 for the wild‐type (Table 1). Similarly, the Fgcap1 mutant caused only limited symptoms on wheat coleoptiles (Fig. 2B). In thick sections of the rachis beneath the spikelets inoculated with PH‐1, extensive infectious hyphae were observed in vascular tissues at 5 days post‐inoculation (dpi). Under the same conditions, the Fgcap1 mutant showed only limited infectious hyphal growth in vascular tissues of the rachis (Fig. 2C). These results suggest that the Fgcap1 mutant is defective in spreading from the inoculation site to nearby spikelets via the rachis, and that FgCAP1 is important for virulence in F. graminearum.
Figure 2.
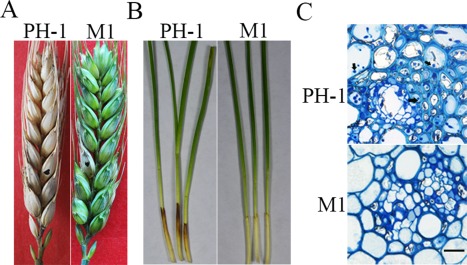
Defects of the Fgcap1 mutant in plant infection. (A) Flowering wheat heads of cultivar Xiaoyan 22 were drop inoculated with conidia from the wild‐type (PH‐1) and Fgcap1 mutant (M1). Spikelets with typical symptoms were photographed at 14 days post‐inoculation (dpi). Black dots mark the inoculated spikelets. (B) Wheat coleoptiles inoculated with PH‐1 and M1 were photographed at 7 dpi. (C) The rachis directly beneath the inoculated spikelets was examined at 5 dpi. Hyphal growth (marked with arrows) was abundant in plant tissues inoculated with PH‐1, but not in samples inoculated with the Fgcap1 mutant M1. Bar, 20 μm.
FgCAP1 plays a critical role in the regulation of DON biosynthesis
Because DON is an important virulence factor, we assayed DON production in infested wheat kernels. The Fgcap1 mutant showed significantly reduced DON production in infested wheat kernels (Table 1). We also assayed DON production with rice grain cultures. In comparison with the wild‐type, DON production was reduced over 10‐fold in the Fgcap1 mutant (Table 1). When assayed by qRT‐PCR with RNA isolated from hyphae collected from 3‐day‐old LTB cultures, the expression levels of TRI5, TRI6, TRI12 and TRI101 were 7.1‐, 2.4‐, 11.1‐ and 6.7‐fold, respectively, higher in the wild‐type than in the Fgcap1 mutant (Fig. 3A). Therefore, FgCAP1 regulates TRI gene expression and DON production in F. graminearum.
Figure 3.
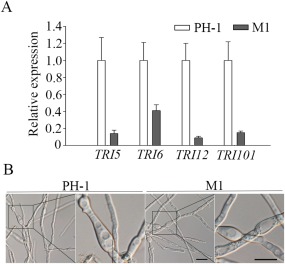
Defects of the Fgcap1 mutant in deoxynivalenol (DON) biosynthesis. (A) The expression levels of TRI5, TRI6, TRI12 and TRI101 in the wild‐type PH‐1 and Fgcap1 mutant M1. The relative expression level of each gene in PH‐1 was arbitrarily set to unity. The means and standard errors were calculated using data from three independent biological replicates. (B) LTB (liquid trichothecene biosynthesis) cultures of PH‐1 and mutant M1 were examined for bulbous structures after incubation for 3 days. Bar, 20 μm.
Because DON production is known to be associated with special cellular differentiation (Jonkers et al., 2012; Menke et al., 2013), the morphology of hyphae harvested from 3‐day‐old LTB cultures was examined. In comparison with PH‐1, no obvious difference was observed in the formation of intercalary, bulbous hyphal structures in the Fgcap1 mutant (Fig. 3B). Therefore, although it is important for DON production, FgCAP1 appears to be dispensable for cellular differentiation associated with DON biosynthesis.
Regulation of DON production by FgCap1 is related to cAMP signalling
As a result of the conserved functions of CAP proteins in cAMP signalling, we assayed the intracellular cAMP level and PKA activity. In vegetative hyphae grown in LTB medium, the intracellular cAMP level in the Fgcap1 mutant was reduced by approximately 70% in comparison with that of the wild‐type strain PH‐1 (Fig. 4A). The Fgcap1 mutant also showed reduced PKA activity (Fig. 4B). These data indicate that FgCAP1 plays an important role in the cAMP‐PKA pathway.
Figure 4.
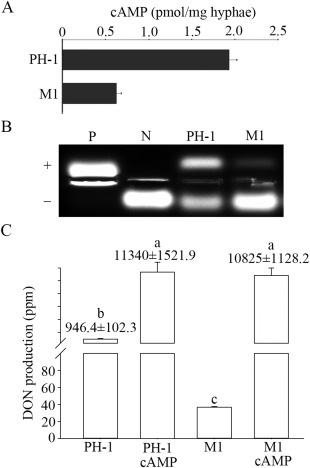
FgCAP1 is involved in cyclic adenosine monophosphate (cAMP) signalling for the regulation of deoxynivalenol (DON) production. (A) Intracellular cAMP level in the wild‐type PH‐1 and Fgcap1 mutant M1. The means and standard errors were calculated using results from three independent biological replicates. (B) Protein kinase A (PKA) activity was assayed with proteins isolated from hyphae of PH‐1 and M1 with the PepTag A1 PKA substrate peptide. Whereas phosphorylated peptides migrated towards the anode (+), non‐phosphorylated peptides migrated towards the cathode (–) on a 0.8% agarose gel. N, non‐phosphorylated sample control; P, phosphorylated sample control. (C) DON production in 7‐day‐old LTB (liquid trichothecene biosynthesis) cultures of PH‐1 and M1 with or without 4 mm cAMP. Different letters indicate statistically significant differences (P = 0.05).
Because the intracellular cAMP level was reduced in the Fgcap1 mutant, we assayed the effect of exogenous cAMP on DON production. As expected, DON production in the wild‐type was significantly increased in the presence of 4 mm cAMP (Fig. 4C). Although the Fgcap1 mutant showed reduced DON production, it still responded to exogenous cAMP and produced similar amounts of DON to the wild‐type in the presence of 4 mm cAMP (Fig. 4C). These results indicate that exogenous cAMP can suppress the defects of the Fgcap1 mutant in DON production. Therefore, the function of FgCAP1 in DON regulation is dependent on the cAMP signalling pathway.
FgCap1 interacts with Fac1 in co‐immunoprecipitation assays
To determine the relationship between FgCap1 and cAMP signalling in F. graminearum, the intracellular region of Fac1 was fused with GFP (FAC1 CT‐GFP) and co‐transformed into PH‐1 with the FgCAP1‐FLAG fusion construct. In western blot analysis with total proteins isolated from the resulting transformant CF1 (Table 2), 61‐kDa and 77‐kDa bands were detected with the anti‐FLAG and anti‐GFP antibodies, respectively (Fig. 5). In the elution from anti‐FLAG M2 beads, the 77‐kDa Fac1CT‐GFP band was detected with an anti‐GFP antibody in the FgCAP1‐FLAG FAC1 CT‐GFP transformant CF1, but not in the transformant expressing only the CAP1‐FLAG construct CC2 (Fig. 5). Therefore, FgCap1 interacts with the C‐terminal region of Fac1 in F. graminearum, further indicating the role of FgCap1 in cAMP signalling.
Table 2.
Wild‐type and mutant strains of Fusarium graminearum used in this study.
| Strain | Genotype description | Reference |
|---|---|---|
| PH‐1 | Wild‐type | Cuomo et al. ( 2007) |
| PH1Δtri6 | tri6 deletion mutant of PH‐1 | Seong et al. ( 2009) |
| M1 | Fgcap1 deletion mutant of PH‐1 | This study |
| C3 | FgCAP1‐complemented transformant of M1 | This study |
| D1 | Complemented transformant of M1 expressing FgCAP1 ΔTri6B allele | This study |
| CC2 | Transformant of PH‐1 expressing FgCap1‐FLAG | This study |
| CF1 | Transformant of PH‐1 expressing FgCap1‐FLAG and FacCT‐GFP | This study |
| RG2 | Transformant of PH‐1 expressing FgCAP1‐RFP and LifeAct‐GFP | This study |
| SG2 | Complemented transformant of M1 expressing FgCAP1 S353A ‐GFP | This study |
| CG5 | FgCAP1‐GFP‐complemented transformant of M1 | This study |
Figure 5.
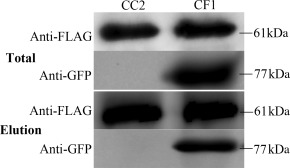
FgCap1 interacts with Fac1 in co‐immunoprecipitation assays. Assays for the interaction of FgCap1 with Fac1. Western blots of total proteins and proteins eluted from anti‐FLAG M2 beads of transformant CC2 (FgCap1‐FLAG) and transformant CF1 (FgCap1‐FLAG and FacCT‐GFP) were detected with the anti‐FLAG or anti‐GFP antibody.
FgCAP1 and TRI6 mediate the feedback regulation of cAMP signalling in the DON‐producing stage
Because exogenous cAMP induced DON production, we measured the intracellular cAMP level in hyphae of PH‐1 harvested from 3‐day‐old YEPD or LTB (DON‐inducing) cultures. The intracellular cAMP level was significantly increased when DON production was induced in LTB cultures in comparison with YEPD cultures (Fig. 6A). When assayed by qRT‐PCR, FgCAP1 expression was also up‐regulated during DON production (Fig. 6B), indicating an association between the intracellular cAMP level and FgCAP1 expression. Interestingly, the tri6 mutant showed no obvious changes in the expression level of FgCAP1 between YEPD and LTB cultures (Fig. 6B). Therefore, as a key regulator of DON biosynthesis, Tri6 may regulate FgCAP1 expression to modulate the intracellular cAMP level as a feedback regulation.
Figure 6.
The putative Tri6‐binding site in the FgCAP1 promoter is important for its function during deoxynivalenol (DON) production. (A) The intracellular cyclic adenosine monophosphate (cAMP) level was assayed using hyphae from the wild‐type strain PH‐1 harvested from 3‐day‐old yeast extract peptone dextrose (YEPD) and LTB (liquid trichothecene biosynthesis) cultures. (B) Expression levels of FgCAP1 in 3‐day‐old YEPD or LTB cultures of PH‐1 and the tri6 mutant. The relative expression level of FgCAP1 in YEPD cultures of PH‐1 was arbitrarily set to unity. Means and standard errors were calculated using results from three independent biological replicates. (C) Three‐day‐old potato dextrose agar (PDA) cultures of PH‐1, Fgcap1 mutant M1 and transformants of M1 expressing the wild‐type FgCAP1 ΔWT (C3) or mutant allele of FgCAP1 ΔTri6B (D1) with deletion of the Tri6‐binding site. (D) Conidia of the same set of strains harvested from 5‐day‐old carboxymethyl cellulose (CMC) cultures. Bar, 20 μm. (E) Typical infected corn silks were photographed at 6 days post‐inoculation (dpi). (F) DON production in 7‐day‐old LTB cultures of the same set of strains.
The promoter of FgCAP1 contains one putative Tri6‐binding site TCACTTCAC (−360 to −351) (Nasmith et al., 2011). To determine its role in FgCAP1 function, we generated a mutant allele of FgCAP1 with deletion of this putative Tri6‐binding site and transformed it into the Fgcap1 mutant. The resulting transformant D1 showed a similar growth rate to the wild‐type (Fig. 6C). It also produced normal conidia (Fig. 6D) and was as virulent as the wild‐type (Fig. 6E). However, deletion of the Tri6‐binding site in FgCAP1 significantly reduced DON production (Fig. 6F), indicating that Tri6 especially regulates FgCAP1 during DON production.
FgCap1 localizes to the apical cortical patches in germ tubes
For complementation assays, we generated the FgCAP1‐GFP construct and transformed it into the Fgcap1 mutant M1. The resulting Fgcap1/FgCAP1‐GFP transformant CG5 was normal in terms of vegetative growth, conidiation, plant infection and DON production, suggesting that the phenotypes of the Fgcap1 mutant could be fully complemented by FgCAP1‐GFP (Fig. 7). When examined by epifluorescence microscopy, GFP signals were observed mainly at the apical region of hyphal tips (Fig. 8A). This localization pattern was similar to that of actin patches in F. graminearum (Li et al., 2015). When FgCAP1‐GFP transformant CG5 was treated by cytochalasin A (CytA), an inhibitor of actin elongation, GFP signals were observed throughout the cytoplasm, instead of mainly at the apical cortical patches (Fig. 8A). These results indicate that FgCap1 is probably associated with the F‐actin cytoskeleton for localization to the cortical regions of hyphal tips.
Figure 7.
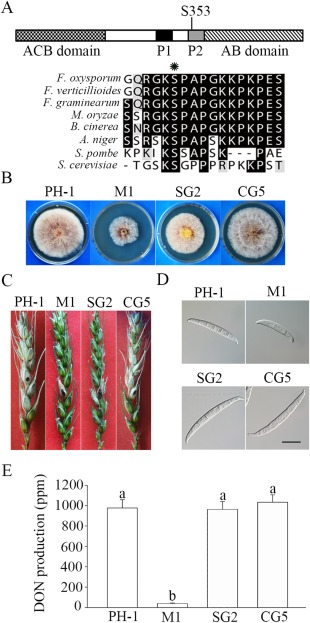
Phosphorylation of FgCap1 at S353 is important for hyphal growth and pathogenesis, but not for conidiogenesis and deoxynivalenol (DON) production. (A) Schematic drawing of FgCap1 and sequence alignment with its orthologues from other fungi surrounding the S353 residue (marked with an asterisk). ACB, adenylyl cyclase‐binding domain; AB, actin‐binding domain; P1 and P2, two proline‐rich regions. (B) Three‐day‐old potato dextrose agar (PDA) cultures of the wild‐type (PH‐1), Fgcap1 mutant (M1), Fgcap1/FgCAP1 S353A‐GFP (SG2) and Fgcap1/FgCAP1‐GFP (CG5) strains. (C) Flowering wheat heads of cultivar Norm were drop inoculated with conidia from the same set of strains. Black dots mark the inoculated spikelets. (D) Conidia harvested from 5‐day‐old carboxymethyl cellulose (CMC) cultures. Bar, 20 μm. (E) DON production in 7‐day‐old LTB (liquid trichothecene biosynthesis) cultures of the same set of strains.
Figure 8.
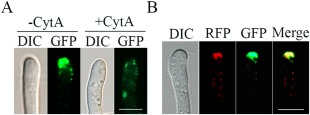
Subcellular localization of FgCap1 and its association with actin. (A) Germlings of transformant CG5 expressing the FgCAP1‐GFP construct treated with or without cytochalasin A (CytA) were examined by differential interference contrast (DIC) and epifluorescence microscopy. Bar, 5 μm. (B) Germlings of transformant RG2 expressing the FgCAP1‐RFP and LifeAct‐GFP constructs were examined by DIC and epifluorescence microscopy. Bar, 5 μm. GFP, green fluorescent protein; RFP, red fluorescent protein.
Because of the localization pattern of FgCap1 and its ABD, we generated the FgCAP1‐RFP construct and co‐transformed it into PH‐1 with the LifeAct‐GFP construct (Riedl et al., 2008). In the resulting transformant RG2, both FgCap1‐RFP and LifeAct‐GFP mainly localized to the cortical regions of hyphal tips, although they did not show identical localization patterns (Fig. 8B).
Phosphorylation of FgCap1 at S353 may play an important role in hyphal growth and pathogenesis
In mouse Cap1, three phosphorylation sites have been identified by mass spectrometric assays (Zhou et al., 2014). To identify the possible phosphorylation sites of FgCap1, we isolated proteins from vegetative hyphae harvested from 12‐h‐old cultures, as described by Pandey et al. (2004). After trypsin digestion, phosphorylated peptides were subjected to mass spectrometric analysis (Gao et al., 2016). In total, we identified 2103 phosphopeptides derived from 1116 F. graminearum proteins from three independent assays (Table S1, see Supporting Information). For FgCap1, one phosphopeptide 350RGKSPAPGKK359, with S353 as the phosphorylation site, was identified. This S353 residue is in the second proline‐rich region (P2) which is adjacent to the C‐terminal ABD (Fig. 7A). Sequence alignment showed that this serine residue is well conserved in Cap1 orthologues from budding yeast and other filamentous fungi (Fig. 7A).
To determine the role of S353 phosphorylation, we generated the FgCAP1 S353A‐GFP allele and transformed it into the Fgcap1 deletion mutant. The resulting Fgcap1/FgCAP1 S353A transformant SG2 grew more rapidly than the original Fgcap1 mutant, but still slightly slower than the Fgcap1/FgCAP1 WT complemented transformant CG5 (Fig. 7B). In infection assays with flowering wheat heads, similar to the Fgcap1 mutant, the Fgcap1/FgCAP1 S353A transformant caused symptoms only on inoculated kernels (Fig. 7C). Interestingly, unlike the Fgcap1 mutant, conidia produced by the Fgcap1/FgCAP1 S353A transformant were normal in size and morphology, similar to those of Fgcap1/FgCAP1 WT (Fig. 7D). The Fgcap1/FgCAP1 S353A transformant was also similar to the wild‐type and complemented transformant CG5 in terms of DON production (Fig. 7E). These data indicate that phosphorylation of FgCap1 at S353 is important for vegetative growth and plant infection, but dispensable for asexual reproduction and DON biosynthesis.
S353 phosphorylation is dispensable for cAMP signalling, but important for infectious growth
Because DON production is not affected by S353A mutation and FgCAP1 is involved in the regulation of DON production via cAMP signalling, we assayed the intracellular cAMP level. In comparison with the original Fgcap1 mutant, the Fgcap1/FgCAP1 S353A transformant SG2 showed increased intracellular cAMP in vegetative hyphae. The intracellular cAMP level was similar between the Fgcap1/FgCAP1 S353A transformant and the wild‐type or complemented transformant CG5 (Fig. 9A). These results indicate that phosphorylation of FgCap1 at S353 is not important for cAMP signalling in vegetative hyphae of F. graminearum.
Figure 9.
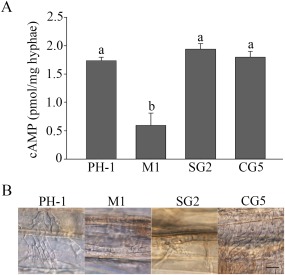
The S353A mutation in FgCap1 affects infectious growth, but not intracellular cyclic adenosine monophosphate (cAMP) levels. (A) Intracellular cAMP level in the wild‐type (PH‐1), Fgcap1 mutant (M1), Fgcap1/FgCAP1 S353A‐GFP (SG2) and Fgcap1/FgCAP1‐GFP (CG5). (B) Infectious hyphae formed by the same set of strains inside wheat coleoptile cells at 48 h post‐inoculation (hpi). Bar, 20 μm.
The Fgcap1/FgCAP1 S353A transformant was defective with regard to spread in wheat heads, but normal in terms of DON production. To further characterize its defects in plant infection, we conducted infection assays with wheat coleoptiles, as described by Liu et al. (2015). The wild‐type and complemented transformant colonized and developed extensive infectious hyphae at 2 dpi. Under the same conditions, the Fgcap1/FgCAP1 S353A transformant, similar to the original Fgcap1 mutant, showed only limited growth of infectious hyphae (Fig. 9B). These results indicate that phosphorylation at S353 plays a critical role in normal infectious growth. In addition to DON production, FgCap1 probably regulates other important pathogenicity factors during plant infection.
Discussion
The cAMP‐PKA signalling pathway is well conserved in eukaryotic organisms and several of its key components, including RAS1, FAC1, PDE1, PDE2, CPK1 and CPK2, have been functionally characterized in F. graminearum (Bluhm et al., 2007; Hu et al., 2014; Jiang et al., 2016b). In the budding yeast, RAS‐dependent activation of adenylate cyclase Cyr1 is required for cAMP synthesis, and the interaction of Cyr1 with the Srv2 CAP protein stimulates its activation (Shima et al., 1997; Zou et al., 2010, Zhou et al., 2012). FgCap1 is orthologous to yeast Srv2 and interacts with Fac1 in F. graminearum. Unlike the fac1 mutant, the Fgcap1 mutant still produces detectable amounts of intracellular cAMP and PKA activity, although at significantly reduced levels in comparison with the wild‐type. Thus, FgCap1 plays an important, but not essential, role in cAMP signalling in F. graminearum. Consistent with this observation, the reduction in growth rate in the Fgcap1 mutant was less than that of the fac1 or cpk1 cpk2 mutant (Hu et al., 2014). Nevertheless, similar to the cpk1 mutant (Hu et al., 2014), the Fgcap1 mutant showed increased tolerance to elevated temperatures during vegetative growth in F. graminearum. The cAMP‐PKA pathway is known to mediate tolerance to elevated temperatures in Fusarium verticillioides and Neurospora crassa (Banno et al., 2005; Choi and Xu, 2010).
In F. graminearum, cAMP signalling plays a critical role in the regulation of DON biosynthesis (Hu et al., 2014; Jiang et al., 2016b). As an activator of Fac1, we found that FgCap1 was important for the expression of TRI genes and that exogenous cAMP fully complemented the defects of the Fgcap1 mutant in DON production. However, FgCap1 was dispensable for the formation of bulbous, swelling hyphal structures that have been observed in DON‐producing cultures (Jonkers et al., 2012). These results suggest that cellular differentiation associated with DON biosynthesis and the expression of TRI genes may be co‐regulated by different mechanisms during DON production. It is likely that FgCAP1 is involved in the regulation of TRI gene expression via cAMP signalling, but not in the cellular differentiation associated with DON production.
The expression of FgCAP1 and the intracellular cAMP level were increased in DON‐inducing cultures of the wild‐type, but not the tri6 mutant. Because of the conserved functions of CAP proteins in cAMP signalling (Zhou et al., 2012), increased expression of FgCAP1 may be responsible for increased cAMP accumulation during DON biosynthesis. The promoter region of FgCAP1 contains one putative Tri6‐binding site, TCACTTCAC, which matches the TCAC N1 TCAC site identified in genes regulated by Tri6 by ChIP‐seq analysis (Nasmith et al., 2011). Deletion of this Tri6‐binding site in FgCAP1 led to a significant reduction in DON production and virulence, but had no effect on vegetative growth. In addition to TRI genes, TRI6 is known to regulate the transcription of a number of genes involved in plant infection, branched‐chain amino acid (BCAA) metabolism and the isoprenoid biosynthetic pathway (Seong et al., 2009; Subramaniam et al., 2015). Because it is in the main TRI gene cluster, TRI6 must have evolved to regulate other genes related to DON biosynthesis and plant infection, including FgCAP1, in F. graminearum. Indeed, the MAC1, CPK1 and CPK2 genes related to cAMP signalling all showed reduced expression levels in the tri6 mutant based on published microarray data (Seong et al., 2009) (www.plexdb.org/). Because cAMP treatment stimulates DON biosynthesis, transcriptional regulation of FgCAP1 by TRI6 may have feedback effects on TRI gene expression by cAMP signalling in F. graminearum.
FgCap1 was mainly localized to the apical cortical patches at hyphal tips. This actin‐like subcellular localization of FgCap1 was disrupted by CytA treatment, indicating the relationship between Cap1 and actin localization. The localization pattern of FgCap1 suggests that it may be functionally related to endocytosis. Apical actin patches may mediate endocytosis in fungi (Berepiki et al., 2011). It has been reported that CAP protein interacts directly with V‐ATPase and acts as a general regulator of endocytosis in Dictyostelium (Sultana et al., 2005). A recent study has shown that Srv2/Cap1 is responsible for efficient endocytosis by aiding initial vesicle invagination and movement (Toshima et al., 2016). Some of the defects observed in the Fgcap1 mutant may be independent of cAMP signalling, but related to the role of FgCAP1 in endocytosis in F. graminearum.
Three phosphorylation sites, S36, S307 and S309, have been identified in the mouse CAP1 protein (Zhou et al., 2014). Although these three phosphorylation sites are conserved in mammalian CAP proteins (Zhou et al., 2014), none are conserved among Cap1 orthologues in filamentous fungi. Nevertheless, the phosphorylation site identified in this study, S353, is conserved in FgCap1 and its orthologues from filamentous ascomycetes. The S353 residue of FgCap1 is in the second proline‐rich domain P2, which is important for the subcellular localization of Cap1 in M. oryzae (Zhou et al., 2012). Interestingly, although phosphorylation of S353 was identified in proteins isolated from vegetative hyphae, the S353A mutation had only a minor effect on the function of FgCap1 during vegetative growth. The Fgcap1/FgCAP1 S353A transformant showed only a slightly reduced growth rate and had similar intracellular cAMP levels to the wild‐type in vegetative hyphae. Expression of FgCAP1 S353A in the Fgcap1 mutant also resulted in the formation of normal conidia and a wild‐type level of DON production. Therefore, phosphorylation of FgCap1 at S353 must be dispensable for its function during conidiogenesis and DON production. However, expression of FgCAP1 S353A in the Fgcap1 mutant failed to suppress its defects in plant infection. The Fgcap1/FgCAP1 S353A transformant was similar to the original Fgcap1 mutant in virulence and was defective in infectious hyphal growth in plant tissues. These results indicate a distinct role of FgCap1 in plant infection, which may be related to the actin cytoskeleton, because the intracellular cAMP level was normal in the Fgcap1/FgCAP1 S353A transformant. Nevertheless, it remains possible that the phosphorylation of FgCap1 is associated with proteins other than Fac1 to regulate the cAMP‐PKA pathway.
It is likely that the function of FgCap1 and its phosphorylation at S353 in cAMP signalling and cytoskeleton reorganization are different between vegetative hyphae and infectious hyphae. In F. graminearum and several other plant‐pathogenic fungi, infectious hyphae have a distinct morphology from vegetative hyphae (Jiang et al., 2016a; Liu et al., 2015; Zhao et al., 2005), which may be related to differences in the functions of FgCap1 in cAMP signalling and cytoskeleton reorganization.
For the three phosphorylation sites of mouse CAP1, the GSK3 kinase is responsible for phosphorylation at S309, but the kinases responsible for phosphorylation at S36 and S307 are not known (Zhou et al., 2014). Because phosphorylation of FgCap1 at S353 is important for infectious growth, it is important to identify the protein kinases responsible for its phosphorylation. In F. graminearum, targeted deletion mutants of all the non‐essential protein kinase genes have been generated (Wang et al., 2011). Phosphoproteomics analysis of these kinase mutants may lead to the identification of the protein kinases responsible for FgCap1 phosphorylation. Prediction with the program KinasePhos showed that S353 matches the consensus phosphorylation site of CDK kinases (Wong et al., 2007). In F. graminearum, there are two Cdc2 orthologues, named Cdc2A and Cdc2B (Liu et al., 2015). Although these two CDC2 orthologues have redundant functions during vegetative growth, CDC2A plays stage‐specific roles in cell cycle regulation during infectious growth (Liu et al., 2015). It is possible that CDC2A is involved in the phosphorylation of FgCap1 in F. graminearum. Nevertheless, the production of conidia with reduced septation (fewer conidium compartments) is a common phenotype between the Fgcap1 and Fggsk3 mutants (Qin et al., 2015). Fggsk3 also shares similar defects in growth, DON production and plant infection with the Fgcap1 mutant. Therefore, it remains possible that FgGSK3 is involved in the activation of FgCAP1 in F. graminearum.
Experimental Procedures
Strains and culture conditions
All the wild‐type and mutant strains of F. graminearum used in this study are listed in Table 2. Potato dextrose agar (PDA) cultures grown at 25 ºC for 3 days were used to assay the growth rate and colony morphology. Conidiation and conidium morphology were assayed with conidia harvested from 5‐day‐old carboxymethyl cellulose (CMC) cultures, as described by Jiang et al. (2016b). For mating assays, aerial hyphae of 7‐day‐old carrot agar cultures were pressed down with 0.1% Tween‐20 and further incubated under black light to induce sexual reproduction (Luo et al., 2014).
Generation of the Fgcap1 deletion mutant
To generate the gene replacement construct for FgCAP1 with the split‐marker approach, the 0.9‐kb upstream and 0.9‐kb downstream flanking sequences were amplified by polymerase chain reaction (PCR) from genomic DNA of PH‐1. The resulting PCR products were connected to the neomycin resistance gene cassette (Itaya et al., 1989) by overlapping PCR and transformed into protoplasts of PH‐1, as described by Hou et al. (2002). For transformant selection, geneticin (Sigma‐Aldrich, St. Louis, MO, USA) was added to a final concentration at 400 μg/mL to both top and bottom agar. Geneticin‐resistant transformants were screened by PCR and confirmed by Southern blot analysis.
Generation of the FgCAP1‐GFP, FgCAP1‐RFP, FgCAP1 ΔTri6B and FgCAP1 S353A‐GFP transformants
For complementation assays, the FgCAP1‐GFP fusion construct was generated with the gap repair approach (Zhou et al., 2011) by co‐transformation of the FgCAP1 gene fragment and XhoI‐digested pDL2 into yeast strain XK1‐25, as described by Zhou et al. (2011). The FgCAP1‐RFP fusion construct was generated with pXY201 (Zhou et al., 2011) using the same approach. The S353A mutation was introduced into FgCAP1 by overlapping PCR with fragments amplified with the primer pairs Cap1‐NF/S353‐SA‐R and S353‐SA‐F/Cap1‐GFP‐R. FgCAP1 S353A was cloned into pFL2 (Zhou et al., 2011) by gap repair to generate the FgCAP1 S353A‐GFP fusion construct. Deletion of the putative Tri6‐binding site in the FgCAP1 promoter was created by overlapping PCR with fragments amplified with the primer pairs Cap1‐NF/S353‐SA‐R and S353‐SA‐F/Cap1‐R. All the resulting fusion constructs and FgCAP1 ΔTri6B allele were recovered from Trp+ yeast transformants, confirmed by sequencing analysis and transformed into protoplasts of the Fgcap1 mutants. Transformants resistant to both hygromycin and bleomycin were screened by PCR and examined for green fluorescent protein (GFP) or red fluorescent protein (RFP) signals, as described by Jiang et al. (2016b). All of the primers used in the construction of these mutant alleles are listed in Table S2 (see Supporting Information).
Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) assays
RNA samples of the wild‐type, tri6 and Fgcap1 mutant were isolated with TRIzol reagent (Invitrogen, Massachusetts, USA) from hyphae harvested from 3‐day‐old yeast extract peptone dextrose (YEPD) and LTB (liquid trichothecene biosynthesis) cultures. To assay the expression levels of FgCAP1 and TRI genes, cDNA was synthesized with the Fermentas First cDNA synthesis kit (Hanover, MD, USA) following the instructions provided by the manufacturer. The iTaq™ Universal SYBR® Green Supermix (Bio‐RAD, USA) was used for qRT‐PCR assays with the CFX96 Real‐Time System, as described previously (Bio‐RAD, California, USA) (Jiang et al., 2016b). The TUB2 β‐tubulin gene of F. graminearum was used as the internal control (Bluhm et al., 2007). The relative expression levels of each gene were calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Plant infection and DON production assays
Conidia harvested from 5‐day‐old CMC cultures were resuspended to 105 spores/mL in sterile distilled water. Wheat heads of cultivar Norm or Xiaoyan 22 were drop inoculated with 10 μL of conidium suspensions. Scab symptoms were examined at 14 dpi and infected wheat kernels were harvested and assayed for DON production, as described by Bluhm et al. (2007). Thick sections of infected rachis tissues, which were fixed, dehydrated and embedded in Spurr resin as described by Kang et al. (2008), were examined for infectious growth in wheat heads. Infection assays with corn silks and wheat coleoptiles were conducted as described previously (Hou et al., 2002; Liu et al., 2015). Infectious hyphae developed in wheat coleoptile cells were examined as described previously (Liu et al., 2015) at 48 h post‐inoculation.
DON production in LTB cultures (Jiang et al., 2016b) was assayed with a competitive enzyme‐linked immunosorbent assay (ELISA)‐based DON detection plate kit (Beacon Analytical Systems, Maine, USA) after incubation at 25 ºC for 1, 3 or 5 days, as described by Gardiner et al. (2009). To stimulate DON production, cAMP (Sigma‐Aldrich) was added to a final concentration of 4 mm to LTB culture, as described by Jiang et al. (2016b). DON production assays with rice grain cultures were conducted as described by Jiang et al. (2015).
Co‐immunoprecipitation assays for the interaction between FgCap1 and Fac1
The FgCAP1‐FLAG and FAC1 CT‐GFP fusion constructs were generated by the yeast gap repair approach (Zhou et al., 2011) and co‐transformed into the wild‐type strain PH‐1. Total proteins were isolated from the resulting transformants, as described by Hou et al. (2015), and the expression of both transforming constructs was verified by western blot analysis with anti‐GFP (Roche, California, USA) and anti‐FLAG (Sigma‐Aldrich) antibodies. Total proteins were isolated from transformants expressing both FgCAP1‐FLAG and FAC1 CT‐GFP constructs and incubated with anti‐Flag M2 beads (Sigma‐Aldrich). Western blots of total proteins and proteins eluted from anti‐Flag M2 beads were detected with anti‐GFP and anti‐FLAG antibodies, as described by Liu et al. (2015).
Assays for intracellular cAMP levels and PKA activities
Hyphae of the wild‐type and Fgcap1 mutant were harvested from 3‐day‐old YEPD and LTB cultures for cAMP assays. The cAMP levels were quantified using the cAMP Biotrak Immuno‐assay System (Amersham Biosciences, New Jersey, USA), as described by Ramanujam and Naqvi (2010). Hyphae of the wild‐type and Fgcap1 mutant cultured in LTB cultures for 3 days were used to extract protein for PKA activity assays. PKA activities were assayed with the PepTag non‐radioactive PKA assay kit (Promega, Wisconsin, USA), as described by Godson et al. (2000).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Table S1 Identification of phosphoprotein and phosphopeptide in Fusarium graminearum.
Table S2 Polymerase chain reaction (PCR) primers used in this study.
Acknowledgements
We thank Dr Chengkang Zhang and Chunlan Wu for assistance with cAMP measurement and plant infection assays. We also thank Drs Yang Li and Xue Zhang for fruitful discussions. This work was supported by the National Basic Research Program of China (2013CB127702), National Natural Science Foundation of China (Nos. 31671981 and 31571953), Zhongying Young Scholars of Northwest A&F University and Young Talent Fund of University Association for Science and Technology in Shaanxi, China (20160203). The authors have no conflicts of interest to declare.
Contributor Information
Jin‐Rong Xu, Email: jinrong@purdue.edu.
Cong Jiang, Email: cjiang@nwafu.edu.cn.
References
- Audenaert, K. , Vanheule, A. , Hofte, M. and Haesaert, G. (2014) Deoxynivalenol: a major player in the multifaceted response of Fusarium to its environment. Toxins, 6, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn, Y.S. and Sundstrom, P. (2001) CAP1, an adenylate cyclase‐associated protein gene, regulates bud‐hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans . J. Bacteriol. 183, 3211–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn, Y.S. , Hicks, J.K. , Giles, S.S. , Cox, G.M. and Heitman, J. (2004) Adenylyl cyclase‐associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP‐protein kinase A cascade. Eukaryot Cell, 3, 1476–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, G.H. , Desjardins, A.E. and Plattner, R.D. (2002) Deoxynivalenol‐nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia, 153, 91–98. [DOI] [PubMed] [Google Scholar]
- Banno, S. , Ochiai, N. , Noguchi, R. , Kimura, M. , Yamaguchi, I. , Kanzaki, S. , Murayama, T. and Fujimura, M. (2005) A catalytic subunit of cyclic AMP‐dependent protein kinase, PKAC‐1, regulates asexual differentiation in Neurospora crassa . Genes Genet. Syst. 80, 25–34. [DOI] [PubMed] [Google Scholar]
- Berepiki, A. , Lichius, A. and Read, N.D. (2011) Actin organization and dynamics in filamentous fungi. Nat. Rev. Microb. 9, 876–887. [DOI] [PubMed] [Google Scholar]
- Bertling, E. , Hotulainen, P. , Mattila, P.K. , Matilainen, T. , Salminen, M. and Lappalainen, P. (2004) Cyclase‐associated protein 1 (CAP1) promotes cofilin‐induced actin dynamics in mammalian nonmuscle cells. Mol. Biol. Cell, 15, 2324–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluhm, B.H. , Zhao, X. , Flaherty, J.E. , Xu, J.R. and Dunkle, L.D. (2007) RAS2 regulates growth and pathogenesis in Fusarium graminearum . Mol. Plant–Microbe Interact. 20, 627–636. [DOI] [PubMed] [Google Scholar]
- Blum, A. , Benfield, A.H. , Stiller, J. , Kazan, K. , Batley, J. and Gardiner, D.M. (2016) High‐throughput FACS‐based mutant screen identifies a gain‐of‐function allele of the Fusarium graminearum adenylyl cyclase causing deoxynivalenol over‐production. Fungal Genet. Biol. 90, 1–11. [DOI] [PubMed] [Google Scholar]
- Boenisch, M.J. and Schafer, W. (2011) Fusarium graminearum forms mycotoxin producing infection structures on wheat. BMC Plant Biol. 11, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormann, J. , Boenisch, M.J. , Bruckner, E. , Firat, D. and Schafer, W. (2014) The adenylyl cyclase plays a regulatory role in the morphogenetic switch from vegetative to pathogenic lifestyle of Fusarium graminearum on wheat. PLoS One, 9, e91135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.W. , Dyer, R.B. , McCormick, S.P. , Kendra, D.F. and Plattner, R.D. (2004) Functional demarcation of the Fusarium core trichothecene gene cluster. Fungal Genet. Biol. 41, 454–462. [DOI] [PubMed] [Google Scholar]
- Catlett, N. , Lee, B.N. , Yoder, O. and Turgeon, B.G. (2003) Split‐marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Newsl. 50, 9–11. [Google Scholar]
- Choi, Y.E. and Xu, J.R. (2010) The cAMP signaling pathway in Fusarium verticillioides is important for conidiation, plant infection, and stress responses but not fumonisin production. Mol. Plant–Microbe Interact. 23, 522–533. [DOI] [PubMed] [Google Scholar]
- Cuomo, C.A. , Guldener, U. , Xu, J.R. , Trail, F. , Turgeon, B.G. , Di Pietro, A. , Walton, J.D. , Ma, L.J. , Baker, S.E. , Rep, M. , Adam, G. , Antoniw, J. , Baldwin, T. , Calvo, S. , Chang, Y.L. , Decaprio, D. , Gale, L.R. , Gnerre, S. , Goswami, R.S. , Hammond‐Kosack, K. , Harris, L.J. , Hilburn, K. , Kennell, J.C. , Kroken, S. , Magnuson, J.K. , Mannhaupt, G. , Mauceli, E. , Mewes, H.W. , Mitterbauer, R. , Muehlbauer, G. , Münsterkötter, M. , Nelson, D. , O'Donnell, K. , Ouellet, T. , Qi, W. , Quesneville, H. , Roncero, M.I. , Seong, K.Y. , Tetko, I.V. , Urban, M. , Waalwijk, C. , Ward, T.J. , Yao, J. , Birren, B.W. and Kistler, H.C. (2007) The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science, 317, 1400–1402. [DOI] [PubMed] [Google Scholar]
- Fedor‐Chaiken, M. , Deschenes, R.J. and Broach, J.R. (1990) SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell, 61, 329–340. [DOI] [PubMed] [Google Scholar]
- Gao, X.L. , Jin, Q.J. , Jiang, C. , Li, Y. , Li, C.H. , Liu, H.Q. , Kang, Z. and Xu, J.R. (2016) FgPrp4 kinase is important for spliceosome B‐complex activation and splicing efficiency in Fusarium graminearum . PLoS Genet. 12, e1005973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, D.M. , Kazan, K. and Manners, J.M. (2009) Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum . Fungal Genet. Biol. 46, 604–613. [DOI] [PubMed] [Google Scholar]
- Godson, C. , Mitchell, S. , Harvey, K. , Petasis, N.A. , Hogg, N. and Brady, H.R. (2000) Cutting edge: lipoxins rapidly stimulate nonphlogistic phagocytosis of apoptotic neutrophils by monocyte‐derived macrophages. J. Immunol. 164, 1663–1667. [DOI] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Guo, L. , Breakspear, A. , Zhao, G.Y. , Gao, L.X. , Kistler, H.C. , Xu, J.R. and Ma, L.J. (2016) Conservation and divergence of the cyclic adenosine monophosphate‐protein kinase A (cAMP‐PKA) pathway in two plant‐pathogenic fungi: Fusarium graminearum and F. verticillioides . Mol. Plant Pathol. 17, 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn, T.M. , Krishna, R. and Proctor, R.H. (1999) Characterization of a transcriptional activator controlling trichothecene toxin biosynthesis. Fungal Genet. Biol. 26, 224–235. [DOI] [PubMed] [Google Scholar]
- Hou, R. , Jiang, C. , Zheng, Q. , Wang, C.F. and Xu, J.R. (2015) The AreA transcription factor mediates the regulation of deoxynivalenol (DON) synthesis by ammonium and cyclic adenosine monophosphate (cAMP) signalling in Fusarium graminearum . Mol. Plant Pathol. 16, 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Z.M. , Xue, C.Y. , Peng, Y.L. , Katan, T. , Kistler, H.C. and Xu, J.R. (2002) A mitogen‐activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant–Microbe Interact. 15, 1119–1127. [DOI] [PubMed] [Google Scholar]
- Hu, S. , Zhou, X.Y. , Gu, X.Y. , Cao, S.L. , Wang, C.F. and Xu, J.R. (2014) The cAMP‐PKA pathway regulates growth, sexual and asexual differentiation, and pathogenesis in Fusarium graminearum . Mol. Plant–Microbe Interact. 27, 557–566. [DOI] [PubMed] [Google Scholar]
- Itaya, M. , Kondo, K. and Tanaka, T. (1989) A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 17, 4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C. , Zhang, S.J. , Zhang, Q. , Tao, Y. , Wang, C.F. and Xu, J.R. (2015) FgSKN7 and FgATF1 have overlapping functions in ascosporogenesis, pathogenesis and stress responses in Fusarium graminearum . Environ. Microbiol. 17, 1245–1260. [DOI] [PubMed] [Google Scholar]
- Jiang, C. , Xu, J.R. and Liu, H. (2016a) Distinct cell cycle regulation during saprophytic and pathogenic growth in fungal pathogens. Curr. Genet. 62, 185–189. [DOI] [PubMed] [Google Scholar]
- Jiang, C. , Zhang, C. , Wu, C. , Sun, P. , Hou, R. , Liu, H. , Wang, C. and Xu, J.R. (2016b) TRI6 and TRI10 play different roles in the regulation of DON production by cAMP signaling in Fusarium graminearum. Environ. Microbiol. 18, 3689–3701. [DOI] [PubMed] [Google Scholar]
- Jonkers, W. , Dong, Y. , Broz, K. and Kistler, H.C. (2012) The Wor1‐like protein Fgp1 regulates pathogenicity, toxin synthesis and reproduction in the phytopathogenic fungus Fusarium graminearum . PLoS Pathog. 8, e1002724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Z. , Buchenauer, H. , Huang, L. , Han, Q. and Zhang, H. (2008) Cytological and immunocytochemical studies on responses of wheat spikes of the resistant Chinese cv. Sumai 3 and the susceptible cv. Xiaoyan 22 to infection by Fusarium graminearum . Eur. J. Plant Pathol. 120, 383–396. [Google Scholar]
- Li, C. , Melesse, M. , Zhang, S. , Hao, C. , Wang, C. , Zhang, H. , Hall, M.C. , and Xu, J.R. (2015) FgCDC14 regulates cytokinesis, morphogenesis, and pathogenesis in Fusarium graminearum . Mol. Microbiol. 98, 770–786. [DOI] [PubMed] [Google Scholar]
- Liu, H.Q. , Zhang, S.J. , Ma, J.W. , Dai, Y.F. , Li, C.H. , Lyu, X.L. , Wang, C. and Xu, J.R. (2015) Two Cdc2 kinase genes with distinct functions in vegetative and infectious hyphae in Fusarium graminearum . PLoS Pathog. 11, e1004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(–Delta Delta C) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Luo, Y. , Zhang, H. , Qi, L. , Zhang, S. , Zhou, X. , Zhang, Y. and Xu, J.R. (2014) FgKin1 kinase localizes to the septal pore and plays a role in hyphal growth, ascospore germination, pathogenesis, and localization of Tub1 beta‐tubulins in Fusarium graminearum . New Phytol. 204, 943–954. [DOI] [PubMed] [Google Scholar]
- Menke, J. , Dong, Y.H. and Kistler, H.C. (2012) Fusarium graminearum Tri12p influences virulence to wheat and trichothecene accumulation. Mol. Plant–Microbe Interact. 25, 1408–1418. [DOI] [PubMed] [Google Scholar]
- Menke, J. , Weber, J. , Broz, K. and Kistler, H.C. (2013) Cellular development associated with induced mycotoxin synthesis in the filamentous fungus Fusarium graminearum . PLoS One, 8, e63077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej, J. , Richard‐Forget, F. and Barreau, C. (2011) The pH regulatory factor Pac1 regulates Tri gene expression and trichothecene production in Fusarium graminearum . Fungal Genet. Biol. 48, 275–284. [DOI] [PubMed] [Google Scholar]
- Nasmith, C.G. , Walkowiak, S. , Wang, L. , Leung, W.W.Y. , Gong, Y.C. , Johnston, A. , Harris, L.J. , Guttman, D.S. and Subramaniam, R. (2011) Tri6 is a global transcription regulator in the phytopathogen Fusarium graminearum . PLoS Pathog. 7, e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, A. , Roca, M.G. , Read, N.D. and Glass, N.L. (2004) Role of a mitogen‐activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa . Eukaryot. Cell, 3, 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, J. , Wang, G. , Jiang, C. , Xu, J.R. and Wang, C. (2015) Fgk3 glycogen synthase kinase is important for development, pathogenesis, and stress responses in Fusarium graminearum . Sci. Rep. 5, 8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero‐Monzon, O. , Jonasson, E.M. , Bertling, E. , Talarico, L. , Chaudhry, F. , Sihvo, M. , Lappalainen, P. and Goode, B.L. (2009) Reconstitution and dissection of the 600‐kDa Srv2/CAP complex: roles for oligomerization and cofilin–actin binding in driving actin turnover. J. Biol. Chem. 284, 10 923–10 934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanujam, R. and Naqvi, N.I. (2010) PdeH, a high‐affinity cAMP phosphodiesterase, is a key regulator of asexual and pathogenic differentiation in Magnaporthe oryzae . PLoS Pathog. 6, e1000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl, J. , Crevenna, A.H. , Kessenbrock, K. , Yu, J.H. , Neukirchen, D. , Bista, M. , Bradke, F. , Jenne, D. , Holak, T.A. , Werb, Z. , Sixt, M. and Wedlich‐Soldner, R. (2008) Lifeact: a versatile marker to visualize F‐actin. Nat. Methods, 5, 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze, L.V. , Beaudry, R.M. , Keller, N.P. and Linz, J.E. (2004) Regulation of aflatoxin synthesis by FadA/cAMP/protein kinase A signaling in Aspergillus parasiticus . Mycopathologia, 158, 219–232. [DOI] [PubMed] [Google Scholar]
- Schmidt‐Heydt, M. , Stoll, D. , Schutz, P. and Geisen, R. (2015) Oxidative stress induces the biosynthesis of citrinin by Penicillium verrucosum at the expense of ochratoxin. Int. J. Food Microbiol. 192, 1–6. [DOI] [PubMed] [Google Scholar]
- Seong, K.Y. , Pasquali, M. , Zhou, X.Y. , Song, J. , Hilburn, K. , McCormick, S. , Dong, Y. , Xu, J.R. and Kistler, H.C. (2009) Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 72, 354–367. [DOI] [PubMed] [Google Scholar]
- Shi, X.O. , Bernhardt, T.G. , Wang, S.M. and Gershon, P.D. (1997) The surface region of the bifunctional vaccinia RNA modifying protein VP39 that interfaces with Poly(A) polymerase is remote from the RNA binding cleft used for its mRNA 5' cap methylation function. J. Biol. Chem. 272, 23 292–23 302. [DOI] [PubMed] [Google Scholar]
- Shima, F. , Yamawaki‐Kataoka, Y. , Yanagihara, C. , Tamada, M. , Okada, T. , Kariya, K. and Kataoka, T. (1997) Effect of association with adenylyl cyclase‐associated protein on the interaction of yeast adenylyl cyclase with Ras protein. Mol. Cell Biol. 17, 1057–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber, C.M. , Lee, W. , Wong, P. , Munsterkotter, M. , Mewes, H.W. , Schmeitzl, C. , Varga, E. , Berthiller, F. , Adam, G. and Güldener, U. (2014) The Fusarium graminearum genome reveals more secondary metabolite gene clusters and hints of horizontal gene transfer. PLoS One, 9, e110311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, R. , Narayanan, S. , Walkowiak, S. , Wang, L. , Joshi, M. , Rocheleau, H. , Ouellet, T. and Harris, L.J. (2015) Leucine metabolism regulates TRI6 expression and affects deoxynivalenol production and virulence in Fusarium graminearum . Mol. Microbiol. 98, 760–769. [DOI] [PubMed] [Google Scholar]
- Sultana, H. , Rivero, F. , Blau‐Wasser, R. , Schwager, S. , Balbo, A. , Bozzaro, S. , Schleicher, M. and Noegel, A.A. (2005) Cyclase‐associated protein is essential for the functioning of the endo‐lysosomal system and provides a link to the actin cytoskeleton. Traffic, 6, 930–946. [DOI] [PubMed] [Google Scholar]
- Takach, J.E. and Gold, S.E. (2010) Identification and characterization of Cap1, the adenylate cyclase‐associated protein (CAP) ortholog in Ustilago maydis . Physiol. Mol. Plant Pathol. 75, 30–37. [Google Scholar]
- Toshima, J.Y. , Horikomi, C. , Okada, A. , Hatori, M.N. , Nagano, M. , Masuda, A. , Yamamoto, W. , Siekhaus, D.E. and Toshima, J. (2016) Srv2/CAP is required for polarized actin cable assembly and patch internalization during clathrin‐mediated endocytosis. J. Cell. Sci. 129, 367–379. [DOI] [PubMed] [Google Scholar]
- Van de Walle, J. , Sergent, T. , Piront, N. , Toussaint, O. , Schneider, Y.J. and Larondelle, Y. (2010) Deoxynivalenol affects in vitro intestinal epithelial cell barrier integrity through inhibition of protein synthesis. Toxicol. Appl. Pharmacol. 245, 291–298. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Zhang, S. , Hou, R. , Zhao, Z. , Zheng, Q. , Xu, Q. , Zheng, D. , Wang, G. , Liu, H. , Gao, X. , Ma, J.W. , Kistler, H.C. , Kang, Z. and Xu, J.R. (2011) Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum . PLoS Pathog. 7, e1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y.H. , Lee, T.Y. , Liang, H.K. , Huang, C.M. , Wang, T.Y. , Yang, Y.H. , Chu, C.H. , Huang, H.D. , Ko, M.T. and Hwang, J.K. (2007) KinasePhos 2.0: a web server for identifying protein kinase‐specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res. 35, W588–W594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Wang, C. , Palmieri, S.J. , Haarer, B.K. and Field, J. (1999) A cytoskeletal localizing domain in the cyclase‐associated protein, CAP/Srv2p, regulates access to a distant SH3‐binding site. J. Biol. Chem. 274, 19 985–19 991. [DOI] [PubMed] [Google Scholar]
- Zhao, X. , Kim, Y. , Park, G. and Xu, J.R. (2005) A mitogen‐activated protein kinase cascade regulating infection‐related morphogenesis in Magnaporthe grisea . Plant Cell, 17, 1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, G.L. , Zhang, H. , Wu, H. , Ghai, P. and Field, J. (2014) Phosphorylation of the cytoskeletal protein CAP1 controls its association with cofilin and actin. J. Cell Sci. 127, 5052–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Li, G. and Xu, J.R. (2011) Efficient approaches for generating GFP fusion and epitope‐tagging constructs in filamentous fungi. Methods Mol. Biol. 722, 199–212. [DOI] [PubMed] [Google Scholar]
- Zhou, X.Y. , Zhang, H.F. , Li, G.T. , Shaw, B. and Xu, J.R. (2012) The cyclase‐associated protein Cap1 is important for proper regulation of infection‐related morphogenesis in Magnaporthe oryzae . PLoS Pathog. 8, e1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, H. , Fang, H.M. , Zhu, Y. and Wang, Y. (2010) Candida albicans Cyr1, Cap1 and G‐actin form a sensor/effector apparatus for activating cAMP synthesis in hyphal growth. Mol. Microbiol. 75, 579–591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Table S1 Identification of phosphoprotein and phosphopeptide in Fusarium graminearum.
Table S2 Polymerase chain reaction (PCR) primers used in this study.


