Figure 6.
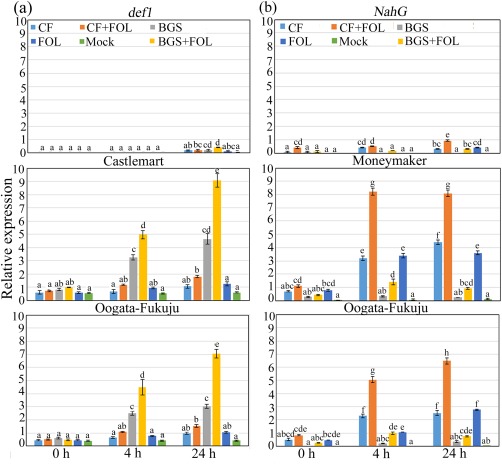
Bar plot of quantitative real‐time reverse transcriptase‐polymerase chain reaction (RT‐qPCR) of the jasmonic acid (JA)‐responsive defensin gene PDF1 and pathogenesis‐related protein 1 acidic (PR1a) gene in 2‐week‐old wild‐type and mutant tomato seedlings treated with different combinations of Trichoderma virens (TriV_JSB100) at different time intervals (0, 4 and 24 h). (a) Relative expression of PDF1 in wild‐type Castlemart and Oogata‐Fukuju seedlings and in JA‐deficient mutant def1 seedlings treated with TriV_JSB100 spores embedded on barley grains (BGS), BGS followed by Fusarium oxysporum f. sp. lycopersici inoculation (BGS + FOL), TriV_JSB100 cell‐free culture filtrate (CF), CF and challenge inoculated with FOL (CF + FOL), FOL (FOL) or sterile distilled water (Mock). (b) Relative expression of PR1a in wild‐type Moneymaker and Oogata‐Fukuju seedlings and in salicylic acid (SA)‐deficient mutant NahG seedlings treated with BGS, BGS + FOL, CF, CF + FOL, FOL and mock. Values are means of four independent replicates (n = 4) ± standard error (SE).Treatment means followed by the same letter(s) within the column are not significantly different according to Tukey's honestly significant difference post hoc test. The β‐actin gene was used as reference. The data presented are from representative experiments that were repeated at least four times with similar results.
