Figure 3.
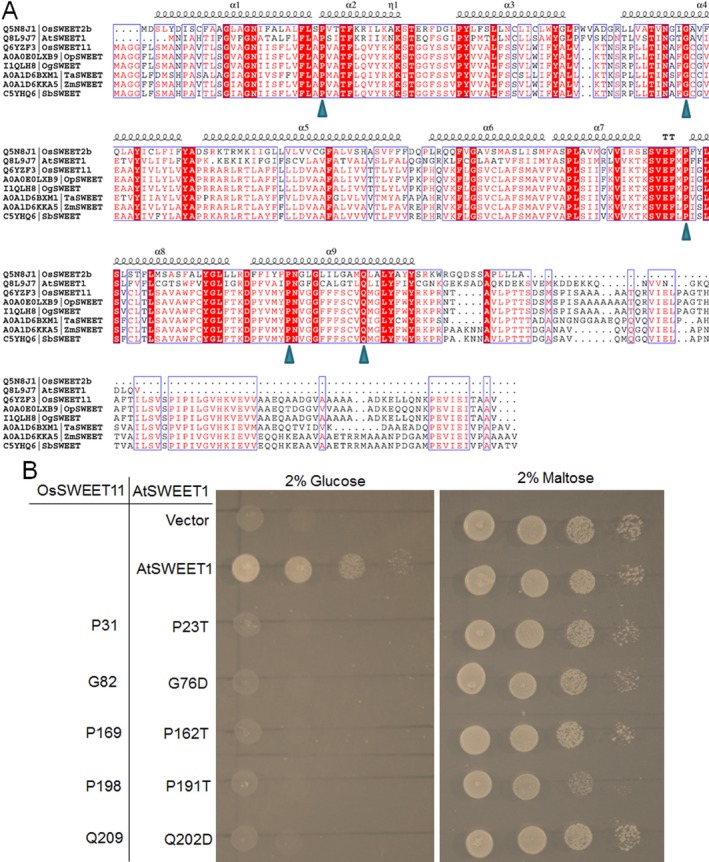
Alignment of amino acid sequences of the selected SWEETs and functional analysis of the conserved residues in glucose transport. (A) Sequence alignment of OsSWEET2b, AtSWEET1a, OsSWEET11, OpSWEET, OgSWEET, TaSWEET, ZmSWEET and SbSWEET. The conserved residues are shown in red boxes. The triangle indicates the residues tested for glucose transport function. (B) Growth assays of mutant AtSWEET1 proteins expressed in EBY4000 yeast strain were performed on YNB (Yeast Nitrogen Base without Amino Acids) medium containing 2% glucose or maltose. AtSWEET1 mutants carrying P23A, G76D, P162A, P191T and Q202D led to the loss of glucose transport activity. Empty (pDRf1) vector and AtSWEET1 were used as negative and positive controls, respectively. The positions of the conserved residues in OsSWEET11 are shown in the left panel. The yeast cells were grown at 28 °C for 3 days.
