Figure 4.
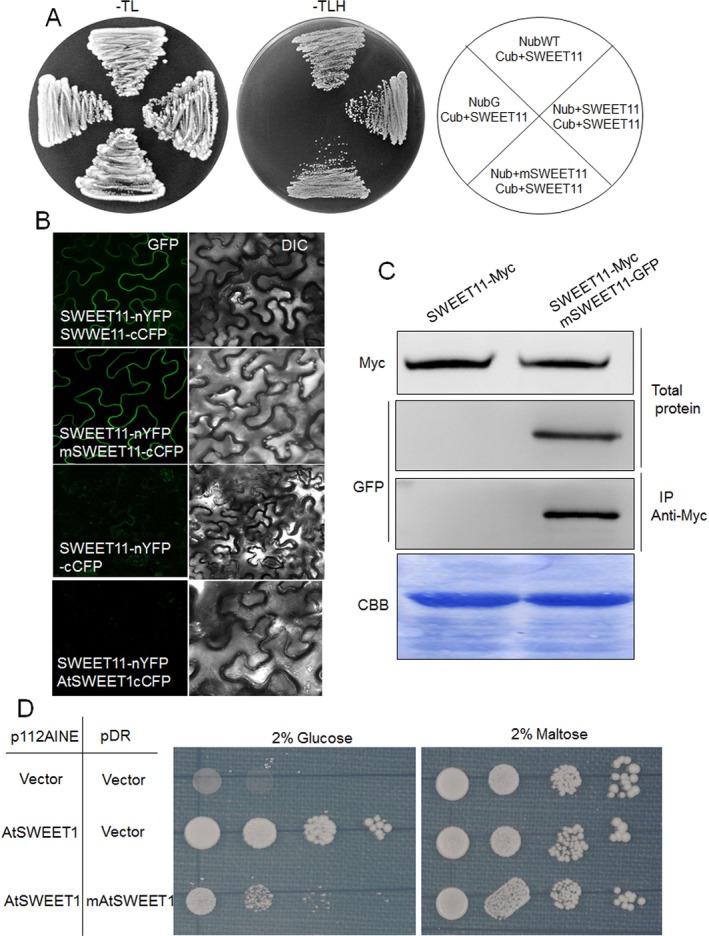
Assessment of the interaction between wild‐type (WT) and mutated OsSWEET11 and inhibition of glucose transport by co‐expression of WT and mutant AtSWEET1 proteins in yeast EBY4000. (A) Interaction between OsSWEET11 and mOsSWEET11 was analysed using a split‐ubiquitin yeast two‐hybrid system. mSWEET11 was cloned into the Nub vector, whereas OsSWEET11 was cloned into the Cub (C‐terminal ubiquitin domain driven by methionine‐repressible MET25 promoter and fused to the artificial PLV transcription factor) or Nub vector. NubWT (Nub) and NubG (N‐terminal ubiquitin domain carrying a glycine mutation) were used as positive and negative controls, respectively. (B) Observation of the yellow fluorescent protein (YFP) fluorescence. Reconstitution of the YFP fluorescence from OsSWEET11‐nYFP + OsSWEET11‐cCFP or OsSWEET11‐nYFP + mOsSWEET11‐cCFP (left, fluorescence channel; right, bright field). Co‐expression of OsSWEET11‐nYFP + cCFP or OsSWEET11‐nYFP + AtSWEET1‐cCFP was used as negative control. Bars, 20 μm. (C) Co‐immunoprecipitation (Co‐IP) was performed to analyse the interaction between OsSWEET11 and mOsSWEET11 in tobacco leaves. OsSWEET11‐Myc or OsSWEET11‐Myc + mOsSWEET11‐GFP was transformed into tobacco leaves using Agrobacterium‐mediated transformation. The total protein and Myc antibody‐immunoprecipitated proteins were analysed using Western blot analysis with either Myc or green fluorescent protein (GFP) antibodies. CBB (Coomassie Brilliant Blue) staining was used as a loading control. (D) The p112AINE empty vector or p112AINE‐SWEET1 construct was co‐transformed with pDRf1 empty vector or pDR‐mAtSWEET1 construct into the yeast strain, EBY4000. Growth assays of yeast cells co‐expressing WT and mutant AtSWEET1 protein, mAtSWEET1, were performed on YNB (Yeast Nitrogen Base without Amino Acids) medium containing 2% glucose or maltose. The yeast cells were grown at 28 °C for 3 days.
