Figure 3.
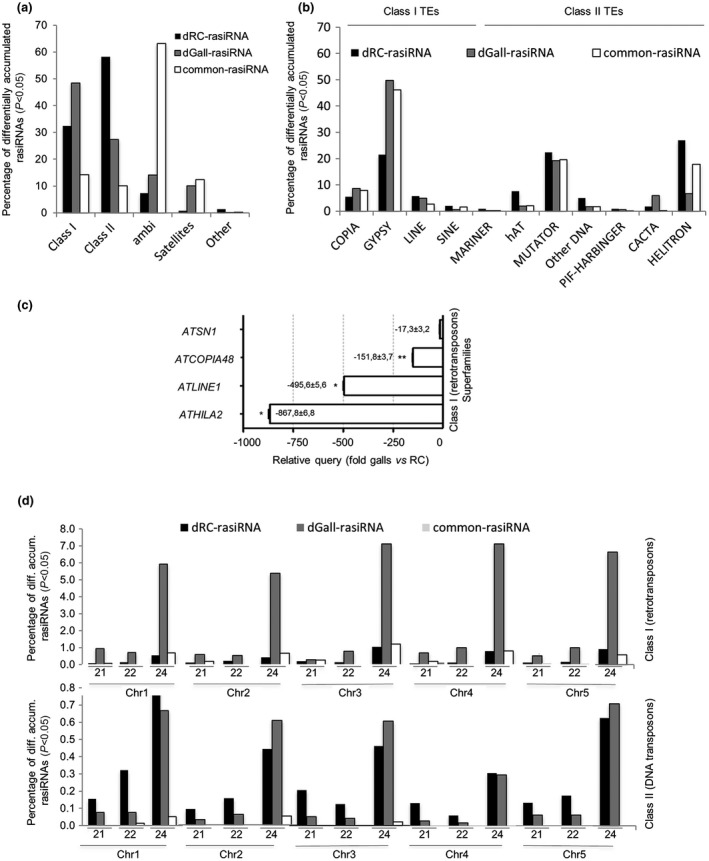
Repression of retrotransposons correlates with the abundance of distinctive gall repeat‐associated small interfering RNAs (rasiRNAs) (24 and 22 nucleotides). (a) Alignment of differentially accumulated rasiRNAs to genome repeat type: class I (retrotransposon), class II (DNA transposon), ambiguous (ambi; siRNAs complementary to more than one transposon element), satellites and other. (b) Percentage of differentially accumulated rasiRNAs that map to different transposon superfamilies also classified as class I and class II transposons. (c) Relative expression levels by quantitative polymerase chain reaction (qPCR) of specific candidates from retrotransposon superfamilies COPIA, GYPSY, LINE and SINE (i.e. ATCOPIA48, ATHILA2, ATLINE1 and ATSN1, respectively) in galls at 3 days post‐infection (dpi) vs. uninfected control roots. Values of two (ATSN1) and four (ATCOPIA48, ATLINE1 and ATHILA2) independent biological replicates, with three technical replicates each, were normalized to Glyceraldehyde‐3‐Phosphate Dehydrogenase (GADPH), used as internal control. Differences from control values were significant at *P < 0.05, **P < 0.01 (two‐tailed t‐test). (d) Mapping of reads normalized to the total amount of 21‐, 22‐ and 24‐nucleotide lengths in each of the five Arabidopsis chromosomes. dGall‐rasiRNA (dark grey), dRC‐rasiRNA (black) and common‐rasiRNA (white). Percentages in (a), (b) and (d) were calculated with respect to the total number of rasiRNAs in each group (dGall‐rasiRNAs, dRC‐rasiRNAs or common‐rasiRNAs).
