Figure 4.
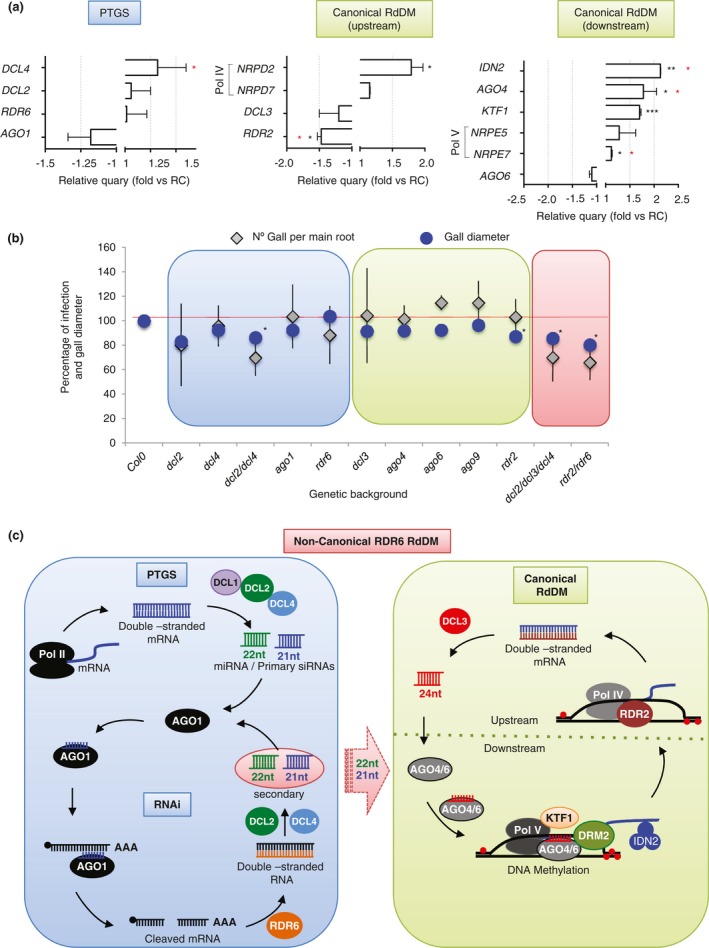
Root‐knot nematode (RKN) infection responses in Arabidopsis mutants of key genes with altered expression in galls impaired in canonical and non‐canonical RDR6‐RdDM pathways. (a) Relative expression levels of canonical, post‐transcriptional gene silencing (PTGS) and non‐canonical RNA‐directed DNA methylation (RdDM) representative genes obtained using quantitative polymerase chain reaction (qPCR) in galls at 3 days post‐infection (dpi) vs. uninfected control roots. The values of two to five independent biological replicates, with three technical replicates each, were normalized to the GADPH internal control. Differences from control values were significant at *P < 0.05, **P < 0.01, ***P < 0.001 (two‐tailed t‐test). A red asterisk indicates P < 0.05 from NEMATIC data (Cabrera et al., 2014). (b) Infection tests with Meloidogyne javanica showing the percentage of galls formed in mutant Arabidopsis lines impaired in repeat‐associated small interfering RNA (rasiRNA) biogenesis compared with the control Col‐0 (grey diamond), as well as the relative gall diameter (n ≥ 27 per line tested) at 14 dpi of the mutant lines relative to that of Col‐0 (blue dots). For infection tests, five to six independent experiments were performed per genetic background with at least 30 plants per independent experiment and transgenic Arabidopsis line. Blue background, mutant Arabidopsis lines from PTGS. Green background, mutant Arabidopsis lines from canonical RdDM pathway. Red background, mutant Arabidopsis lines from PTGS and canonical RdDM pathways. (c) Schematic diagram of non‐canonical RDR6‐RdDM pathway that connects PTGS and canonical RdDM through 21‐ and 22‐nucleotide rasiRNAs in Arabidopsis plants, based on Cuerda‐Gil and Slotkin (2016). PTGS: polymerase II (Pol II) transcription of transposable elements (TEs) or microRNA precursors generates primary small RNAs in an RNA‐dependent RNA polymerase (RDR)‐independent manner after cleavage by DICER‐LIKE (DCLs). Then, RDR6 generates double‐stranded RNAs (dsRNAs) that are triggered by DCL2 and DCL4 to produce secondary rasiRNAs. These can target additional copies of mRNA for the cleavage and production of more rasiRNAs through RNA interference (RNAi). Canonical RdDM: Pol IV and RDR2 produce dsRNA cleaved by DCL3. Then, 24‐nucleotide siRNAs are loaded into AGO4 and AGO6 to chromatin‐bound transcripts produced by Pol V (Upstream). After AGO4/6 interacts with Pol V transcripts, many other proteins will be recruited (IDN2, KTF1, etc.) to finally methylate the DNA through DNA methyltransferase DRM2 (Downstream). Non‐canonical RDR6‐RdDM: 21−22‐nucleotide rasiRNAs produced from Pol II–RDR6‐derived TE mRNAs are loaded into AGO6, which interacts with its target loci through a Pol V scaffolding transcript. [Colour figure can be viewed at wileyonlinelibrary.com]
