Figure 3.
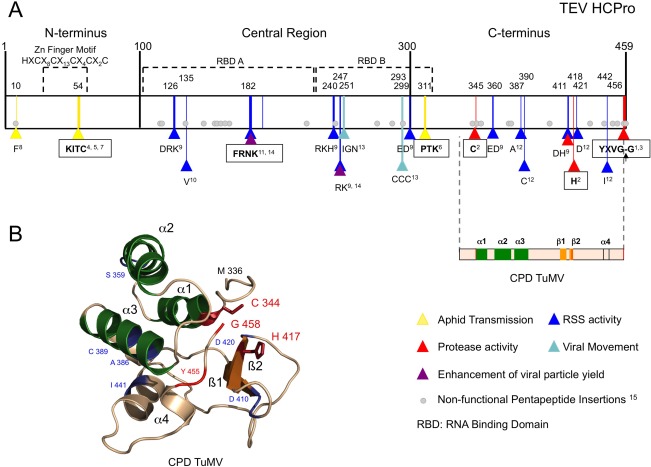
Helper component proteinase (HCPro) structural and functional features. (A) Schematic representation of a representative potyviral HCPro (from Tobacco etch virus, TEV) divided into three main regions. The best‐characterized motifs are shown in squares. Amino acids relevant for a given function, which are conserved at least among the Potyvirus genus, are marked with triangles at their corresponding positions. Amino acids relevant for viral movement (marked in light blue) were described before the characterization of HCPro RNA silencing suppression activity; therefore, their real role might be misassigned. Pentapeptide insertions that render Plum pox virus (PPV) HCPro poorly functional or non‐functional as an RNA silencing suppressor (RSS) are depicted as grey circles at the equivalent TEV HCPro positions. A two‐dimensional representation of the Turnip mosaic virus HCPro structure solved by Guo et al. (2011) encompasses the equivalent C‐terminal region of TEV HCPro. Superscript numbers indicate the following references: 1Carrington et al. (1989a); 2Oh and Carrington (1989); 3Carrington and Herndon (1992); 4Atreya et al. (1992); 5Atreya and Pirone (1993); 6Huet et al. (1994); 7Dolja et al. (1993); 8Blanc et al. (1998); 9Kasschau and Carrington (2001); 10González‐Jara et al. (2005); 11Shiboleth et al. (2007); 12Torres‐Barceló et al. (2008); 13Cronin et al. (1995); 14Valli et al. (2014); 15Varrelmann et al. (2007). (B) Crystal structure of the cysteine protease domain of Turnip mosaic virus HCPro (Guo et al., 2011; PDB code 3RNV). The corresponding L and R domains of papain‐like proteases would be represented by the α‐helices shown in green and the β‐sheets shown in orange, respectively. Those amino acids highlighted in (A) are also indicated in (B).
