Figure 7.
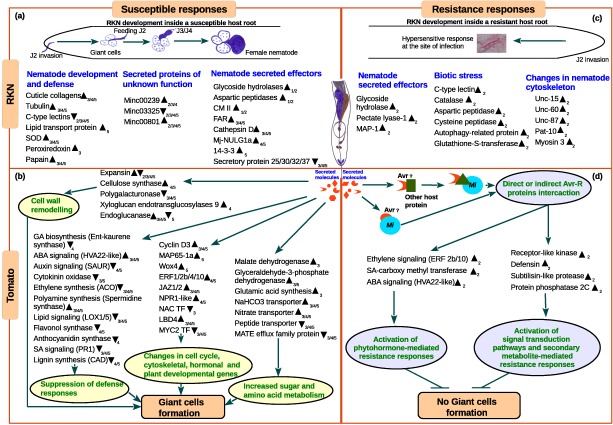
A model depicting the complex changes in the biochemical processes in both tomato and root‐knot nematode (RKN) during susceptible and resistant interactions based on the summary of transcriptome data. (a) Changes in the expression of RKN genes during the susceptible response. (b) Changes in the expression of tomato genes during the susceptible response. (c) Changes in the expression of RKN genes during the resistance response. (d) Changes in the expression of tomato genes during the resistance response. The up‐arrowhead indicates up‐regulated genes, down‐arrowhead indicates down‐regulated genes and numerical subscript in the arrowhead indicates the specific stage in which expression was observed. Host metabolic processes that are modulated on RKN infection are highlighted in coloured circles. ABA, abscisic acid; ACO, 1‐aminocyclopropane‐1‐carboxylic acid oxidase; Avr, avirulence protein; CAD, cinnamoyl alcohol dehydrogenase; CM II, chorismate mutase type II; ERF, ethylene‐responsive transcription factor; FAR, nematode fatty acid retinol‐binding protein; GA, gibberellins; LBD, lateral organ body domain‐containing transcription factor; LOX, lipoxygenase; MAP‐1, Meloidogyne avirulence protein; MATE, multidrug and toxic compound extrusion; NAC TF, no apical meristem‐containing transcription factor; MAP65‐1a, microtubule‐associated protein 65‐1a; pat, paralysed arrest two‐fold; PR1, pathogenesis‐related protein 1; SA, salicylic acid; SOD, superoxide dismutase; unc, uncoordinated; Wox4, WUS homeobox‐containing gene.
