Figure 5.
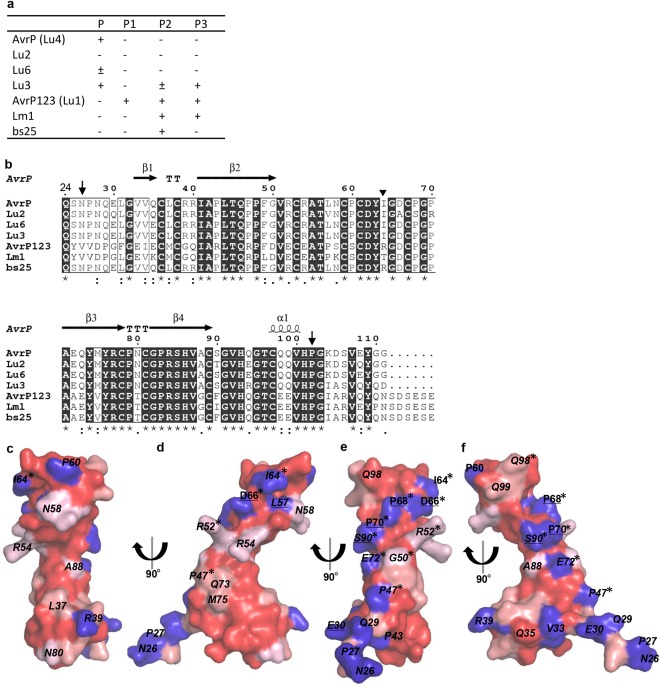
Polymorphic residues on the surface of AvrP variants. (a) The recognition spectra of AvrP variants by P family resistance proteins (Dodds and Thrall, 2009). The AvrP variants were expressed in flax leaves containing the flax P family resistance genes by Agrobacterium infiltration. Plants were scored 12 days after infiltration. ‘+’ indicates that cell death was induced, and ‘–’ indicates that no response was induced. (b) Sequence alignment of AvrP variants. The sequences of mature proteins were aligned using Clustal Omega (Sievers et al., 2011). Residue conservation is indicated below the alignment by an asterisk (fully conserved), a colon (strongly similar) and a period (weakly similar), whereas blank means that there is no similarity. The secondary structure elements of AvrP are shown on top of the alignment and the region included in the structure is located between the downward arrows. ‘T’ corresponds to a β‐turn. The inverted triangle indicates the point at which the bs25 sequence changes from AvrP to the AvrP123 sequence. (c–f) Conservation mapping on the surface of the AvrP structure. The degree of conservation, from strong to weak, is indicated by dark to light red. Non‐conserved residues are shown in blue. Surface residues that are polymorphic between AvrP and Lu2, and AvrP and AvrP123, are underlined and italicized, respectively. Residues targeted for mutation are marked with an asterisk.
