Figure 1.
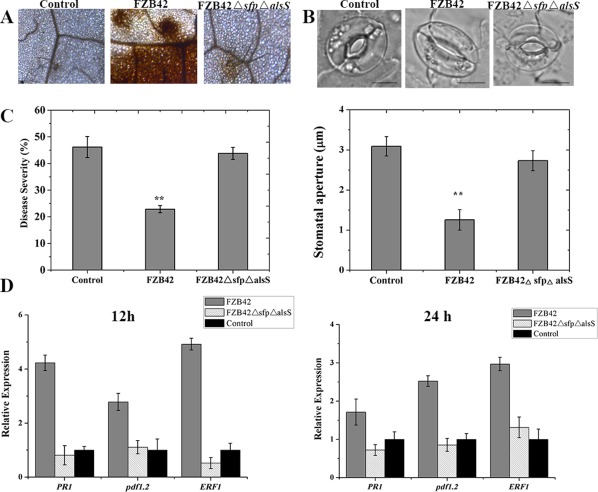
Hydrogen peroxide (A), stomatal aperture (B), disease severity (C) and expression of disease resistance‐related genes (D) in Arabidopsis Col‐0 leaves after inoculation with Bacillus amyloliquefaciens FZB42, the FZB42ΔsfpΔalsS mutant and control. (A) Detection of hydrogen peroxide in Arabidopsis leaves after inoculation with B. amyloliquefaciens FZB42, the FZB42ΔsfpΔalsS mutant and control by 3,3′‐diaminobenzidine (DAB) staining. Brown precipitates represent hydrogen peroxide generation. (B) Stomatal apertures of Arabidopsis leaves after inoculation with B. amyloliquefaciens FZB42, the FZB42ΔsfpΔalsS mutant and control. Scale bars indicate 10 μm. Values represent the means of 50 random selected stomata. All experiments were repeated three times. **Highly significant difference (P < 0.01). (C) Disease severity of Arabidopsis against Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) after inoculation with B. amyloliquefaciens FZB42, the FZB42ΔsfpΔalsS mutant and control. Four‐week‐old Arabidopsis Col‐0 plants were inoculated with cell suspensions of FZB42, FZB42ΔsfpΔalsS and water control. Five days later, leaves were sprayed with a cell suspension of Pst DC3000 at 108 colony‐forming units (CFU)/mL. The disease severity was determined according to the disease index measured at 7 days post‐inoculation (dpi). Each treatment had 12 plants and the experiment was repeated three times. (D) Expression levels of disease resistance‐related genes in Arabidopsis leaves after inoculation with B. amyloliquefaciens FZB42, the FZB42ΔsfpΔalsS mutant and control for 12 and 24 h. Pathogenesis‐related protein (PR1), plant defence factor 1.2 (PDF 1.2) and ethylene response factor 1 (ERF1) genes, involved in the salicylic acid‐, jasmonic acid‐ and ethylene‐dependent defence signalling pathways in Arabidopsis. Data are expressed as the relative expression of the respective mRNAs normalized to the endogenous actin 2. Error bars represent significant differences according to Fisher's least‐significant difference test (P = 0.05) using SPSS software. The experiment was repeated three times.
