Figure 5.
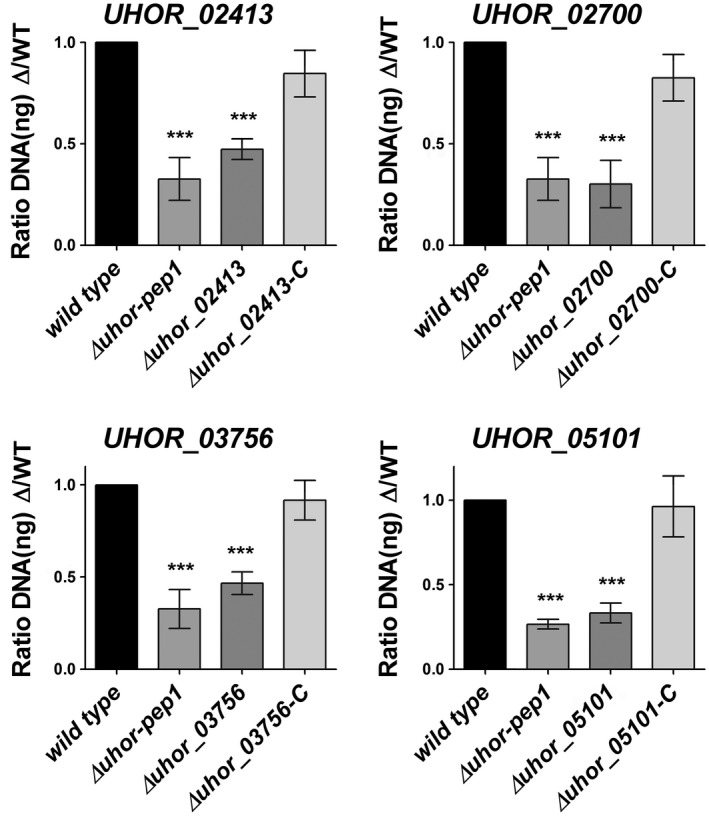
Gene deletion and complementation of Ustilago hordei effector genes. The virulence of the U. hordei wild‐type (WT), four effector mutants and four complementation strains was assessed by the quantification of fungal biomass at 6 days post‐inoculation (dpi). The U. hordei Pep1 deletion mutant was used as a positive control for virulence reduction. The infected barley leaves at 6 dpi were harvested and analysed via quantitative polymerase chain reaction (qPCR) using genomic DNA and the Ppi1 genes from U. hordei as a standard. The fungal biomass was deduced from a standard curve. A one‐way analysis of variance (ANOVA) was performed and followed by a Bonferroni test. Significant differences are indicated with asterisks (***P < 0.001). Error bars represent the standard deviation of at least three biological repeats.
