Figure 3.
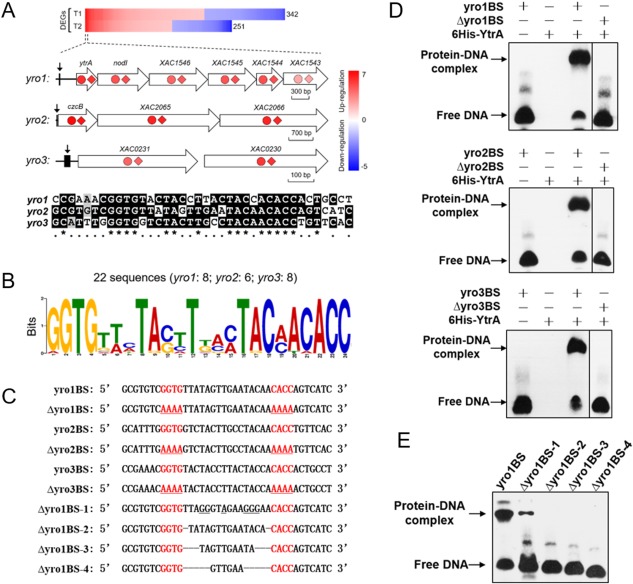
YtrA regulates yro1, yro2 and yro3 regulons and binds to the GGTG‐N16‐CACC palindromic motif. (A) Combined chromatin immunoprecipitation‐exonuclease (ChIP‐exo) and microarray data analysis revealed three operons directly regulated by YtrA. The genes identified by ChIP‐exo were present in the top 30 differentially expressed genes (DEGs) of the microarray data at both T1 and T2 time points. Red represents up‐regulation and blue represents down‐regulation. The fold changes of genes in the three operons at T1 and T2 are illustrated by coloured circles and diamonds, respectively. The YtrA binding positions are marked with arrows. Sequence alignment of YtrA binding sites is shown below the schematic graph. A 38‐bp sequence between two peaks generated by ChIP‐exo was extracted from each regulon. Conserved nucleotides are marked with asterisks. (B) Sequence logo representation of the YtrA binding motif. In total, 22 sequences were submitted to MEME for motif identification. Sequences are listed in Table S2. (C) Sequences of probes used in electrophoretic mobility shift assay (EMSA). Underlined nucleotides indicate the mutations. (D) YtrA binds to motifs located at promoter regions of yro1, yro2 and yro3, respectively. EMSA experiments were conducted in the presence of purified 6His‐YtrA and the corresponding probes listed in (C). (E) Influences of mutated nucleotides on YtrA–DNA interaction. The sequences of the indicated probes are listed in (C). Straight lines indicate deletion of the corresponding nucleotides.
