Figure 3.
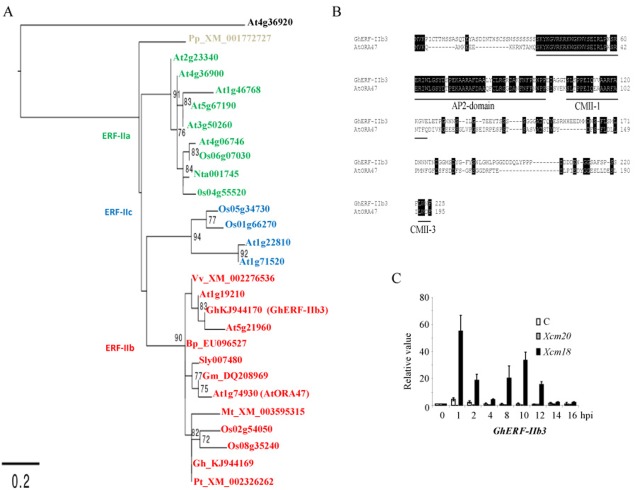
Identification of an ethylene‐response factor (ERF)‐related transcription factor induced during Xanthomonas citri pv. malvacearum (Xcm)‐induced hypersensitive reaction (HR) in cotton plants. (A) A maximum likelihood tree representing relationships among ERF proteins from Arabidopsis thaliana (At), Oryza sativa (Os), Medicago truncatula (Mt), Solanum lycopersicum (Sly), Nicotiana tabacum (Nt), Glycine max (Gm), Vitis vinifera (Vv), Populus trichocarpa (Pt), Broussonetia papyrifera (Bp), Physcomitrella patens (Pp) and Gossypium hirsutum (Gh) plants is rooted to the APETALA2 (AP2) domain R1 (At4g36920). Bootstrap values from 100 replicates were used to assess branch support. Bootstrap values over 70 are shown. The classification by Nakano et al. (2006) is indicated by coloured accession numbers. (B) Alignment of the Gossypium hirsutum GhERF‐IIb3 with the Arabidopsis ERF transcription factor AtORA47. Underlined sequences represent the AP2 domain and the two ERF group IIb‐specific conserved motifs CMII‐1 and CMII‐3. Strictly conserved residues are highlighted in black. Numbers on the right of the alignment indicate the amino acid position for ERF transcription factors. (C) Expression levels of GhERF‐IIb3 in response to Xanthomonas virulent (Xcm20) and avirulent (Xcm18) strains, measured by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The results are averages; error bars indicate standard deviation (n > 12). These experiments were repeated at least three times with similar results.
