Figure 2.
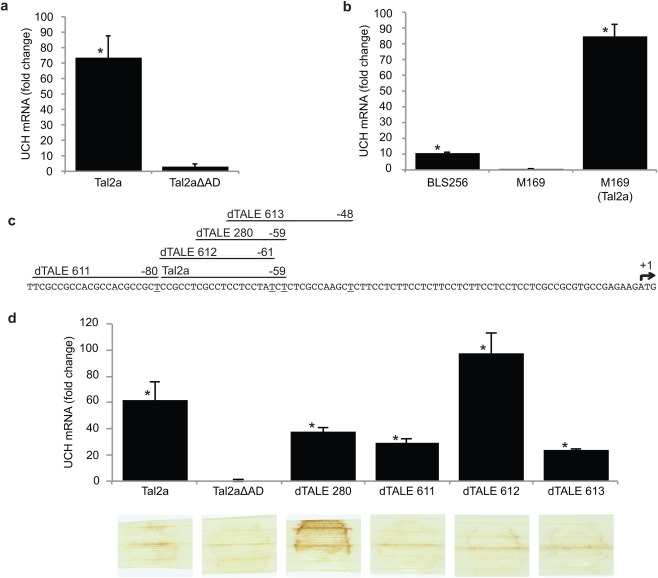
A ubiquitin carboxy‐terminal hydrolase (UCH) gene is activated by Tal2a, but is insufficient for the hypersensitive reaction (HR). (a) Quantitative real‐time reverse transcription‐polymerase chain reaction (qPCR) assay of expression of the UCH gene in response to Tal2a or Tal2aΔAD delivered by Xanthomonas oryzae pv. oryzae PXO99A (Xoo), relative to mock‐inoculated tissue. Tal2a was expressed from plasmid pKEB31. Tissue was sampled at 48 h after syringe infiltration of bacterial suspensions. Each value shown is the average of two technical replicates of three biological replicates. Capped vertical lines show the standard deviation. The relative fold change was calculated by the 2–ΔΔCt method. Asterisks indicate values significantly different (P ≤ 0.05) from mock as determined by two‐tailed, heteroscedastic t‐tests. This experiment was repeated three times with similar results. (b) qPCR assay of UCH expression as in (a) in response to wild‐type X. oryzae pv. oryzicola BLS256, tal2a mutant M169 and M169 carrying the tal2a gene in plasmid pKEB31. Independent experiments were repeated twice with similar results. (c) The 101 nucleotides of the UCH gene upstream of the translational start site showing the predicted effector‐binding element (EBE) for Tal2a and EBEs targeted by each of four designer transcription activator‐like effectors (dTALEs) designed to activate transcription of the gene. Numbers indicate the position of the last (3′) nucleotide of each EBE (underlined) relative to the translational start (+1, arrow). (d) qPCR results as in (a) showing the activation of the UCH gene by each of the four dTALEs delivered by Xoo and, below, the response of rice leaves (as in Fig. 1) to each. Each dTALE activated the UCH gene, but, of these, only dTALE 280, the EBE of which is contained entirely within the Tal2a EBE, triggered an HR, and this HR was stronger than that induced by Tal2a. Experiments were repeated at least twice with similar results.
